Abstract
Mucopolysaccharidoses (MPS) are lysosomal diseases caused by deficiencies in lysosomal enzymes involved in the degradation of glycosaminoglycans (GAGs). Sensorineural hearing impairment is a common feature in MPS patients, but there is no consensus on its etiology. For this reason, we aimed to identify genes and pathways related to hearing loss and to correlate them with gene expression data in MPS. We used HPO and Disgenet to identify candidate genes. We constructed the network with string and Cytoscape, and hub genes were identified in Cytohubba. Expression data were obtained from the MPSBase website. We found the NDUFA gene family as the major hub genes and 114 enriched pathways related to hearing loss. These genes and biological pathways may serve as potential candidates for clinical studies to better understand hearing impairment mechanisms in lysosomal storage diseases like mucopolysaccharidosis.
Keywords:
Ear development; hearing impairment; network analysis; lysosomal storage diseases
Introduction
Mucopolysaccharidoses (MPS) are inborn errors of metabolism characterized by deficiencies in lysosomal enzymes involved in the degradation of glycosaminoglycans (GAGs) which are constituents of the extracellular matrix. These multi-systemic diseases are classified into types based on the differences between the enzyme deficiency and the accumulated GAGs. The ear-nose-throat (ENT) manifestations are common in MPS patients with recurring ear infections and frequently present hearing loss [11. Broek BTA, Smit AL, Boelens JJ, Hasselt PM. Hearing loss in patients with mucopolysaccharidoses ‐1 and ‐6 after hematopoietic cell transplantation: a longitudinal analysis. J Inherit Metab Dis. 2020;43(6):1279-1287. doi:10.1002/jimd.12277.
https://doi.org/10.1002/jimd.12277...
]. Hearing impairment significantly impacts patients’ quality of life, affecting speech and language development [22. Nagao K, Morlet T, Haley E, et al. Neurophysiology of hearing in patients with mucopolysaccharidosis type IV. Mol Genet Metab. 2018;123(4):472-478. doi:10.1016/j.ymgme.2018.02.002.
https://doi.org/10.1016/j.ymgme.2018.02....
].
Hearing loss is common in almost all types of MPS, except in MPS IVB and MPS IX, and can be classified as conductive, sensorineural, or mixed. Each MPS type has a specific type of hearing loss [33. Wolfberg J, Chintalapati K, Tomatsu S, Nagao K. Hearing loss in mucopolysaccharidoses: current knowledge and future directions. Diagnostics (Basel). 2020;10(8):554. doi:10.3390/diagnostics10080554.
https://doi.org/10.3390/diagnostics10080...
]. The conductive occurs when sound conduction is impeded through the ear [44. Isaacson JE, Vora NM. Differential diagnosis and treatment of hearing loss. Am Fam Physician. 2003;68(6):1125-1132.] due to ossicular chain deformities or disruption, or seromucous otitis [11. Broek BTA, Smit AL, Boelens JJ, Hasselt PM. Hearing loss in patients with mucopolysaccharidoses ‐1 and ‐6 after hematopoietic cell transplantation: a longitudinal analysis. J Inherit Metab Dis. 2020;43(6):1279-1287. doi:10.1002/jimd.12277.
https://doi.org/10.1002/jimd.12277...
]. Sensorineural occurs due to problems in the cochlea or the neural pathway to the auditory cortex problems [44. Isaacson JE, Vora NM. Differential diagnosis and treatment of hearing loss. Am Fam Physician. 2003;68(6):1125-1132.]. Sensorineural hearing impairment is a common feature in MPS patients, but there is no consensus on its etiology [33. Wolfberg J, Chintalapati K, Tomatsu S, Nagao K. Hearing loss in mucopolysaccharidoses: current knowledge and future directions. Diagnostics (Basel). 2020;10(8):554. doi:10.3390/diagnostics10080554.
https://doi.org/10.3390/diagnostics10080...
]. The mixed type is the concomitant conductive and sensorineural loss.
Although hearing impairment is a common feature in MPS patients, the available treatments do not significantly improve hearing loss [11. Broek BTA, Smit AL, Boelens JJ, Hasselt PM. Hearing loss in patients with mucopolysaccharidoses ‐1 and ‐6 after hematopoietic cell transplantation: a longitudinal analysis. J Inherit Metab Dis. 2020;43(6):1279-1287. doi:10.1002/jimd.12277.
https://doi.org/10.1002/jimd.12277...
,33. Wolfberg J, Chintalapati K, Tomatsu S, Nagao K. Hearing loss in mucopolysaccharidoses: current knowledge and future directions. Diagnostics (Basel). 2020;10(8):554. doi:10.3390/diagnostics10080554.
https://doi.org/10.3390/diagnostics10080...
]. The mechanisms of hearing loss in MPS remain to be elucidated. Studying and identifying pathways related to hearing can provide helpful information that can be used to develop new therapies.
Methods
Gene Candidate Analysis
We searched for the terms “hearing loss” in the Human Phenotype Ontology - HPO [55. Köhler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021;49(D1):D1207-D1217. doi:10.1093/nar/gkaa1043.
https://doi.org/10.1093/nar/gkaa1043...
] and in the Disgenet databases [66. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845-D855. doi:10.1093/nar/gkz1021.
https://doi.org/10.1093/nar/gkz1021...
]. The HPO database provides a standardized vocabulary of phenotypic abnormalities encountered in human diseases, and their related genes. The Disgenet database has a similar approach, but the database integrates information of human gene-disease association and variant-disease associations from various external databases with information about Mendelian, complex and environmental diseases.
The related terms found in the HPO and Disgenet are shown in Table 1. Then, we used the unique genes between the two databases in the following analysis steps. The flowchart of the methodology is shown in Figure 1.
Network Analysis
A gene network was constructed with the selected candidate genes in the STRING database v.11.0 [77. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. doi:10.1093/nar/gky1131.
https://doi.org/10.1093/nar/gky1131...
]. We used high confidence network scores (0.07) to obtain only experimental data, and text mining interactions were excluded from the analysis. Only the query proteins were considered, without first and second shell interactors. The analyses were performed in Cytoscape v.3.8, with curated plugins [88. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. doi:10.1101/gr.1239303.
https://doi.org/10.1101/gr.1239303...
]. To identify the hub genes, we used Cytohubba v.0.1 with the local based method Maximal Clique Centrality, MCC [99. Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(4 Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11.
https://doi.org/10.1186/1752-0509-8-S4-S...
].
Gene Set Enrichment and Expression Analysis
The functional enrichment was quantitatively assessed (p-value) using a hypergeometric distribution. Multiple test correction was also implemented by applying the FDR at a significance level of p<0.05. We used the Biological Network Gene Ontology (BiNGO) plugin v.3.0.4 [1010. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448-3449. doi:10.1093/bioinformatics/bti551.
https://doi.org/10.1093/bioinformatics/b...
] to identify the biological processes (BP), molecular function (MF), and cellular component pathways. To determine the KEGG [1111. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353-D361. doi:10.1093/nar/gkw1092.
https://doi.org/10.1093/nar/gkw1092...
] pathways, we used the pathfindR package [1212. Ulgen E, Ozisik O, Sezerman OU. pathfindR: An R Package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet. 2019;10:858. doi:10.3389/fgene.2019.00858.
https://doi.org/10.3389/fgene.2019.00858...
] in the R environment [1313. R Core Team. R: A Language and Environment for Statistical Computing [Computer Software]. Vienna, Austria: R Foundation for Statistical Computing; 2017. https://www.R-project.org/.
https://www.R-project.org/...
]. To evaluate the expression of the hub genes, we searched for datasets in the MPSBase [1414. Soares LDF, Silva GCV, Kubaski F, Giugliani R, Matte U. MPSBase: Comprehensive repository of differentially expressed genes for mucopolysaccharidoses. Mol Genet Metab. 2021;133(4):372-377. doi:10.1016/j.ymgme.2021.06.004.
https://doi.org/10.1016/j.ymgme.2021.06....
]. We also selected the most frequent genes which appear in the pathways to evaluate their expression in the available transcriptomic MPS datasets, which were obtained from human IPS and Hela cells.
Results
In HPO, we found 1393 genes, and in Disgenet 1078 (Figure 1). In total, 1679 unique genes were present in either bank. After removing genes without any connections by String, 1617 remained. The Cytoscape network was composed of 827 nodes (genes), and 3777 edges (number of interactions between the genes).
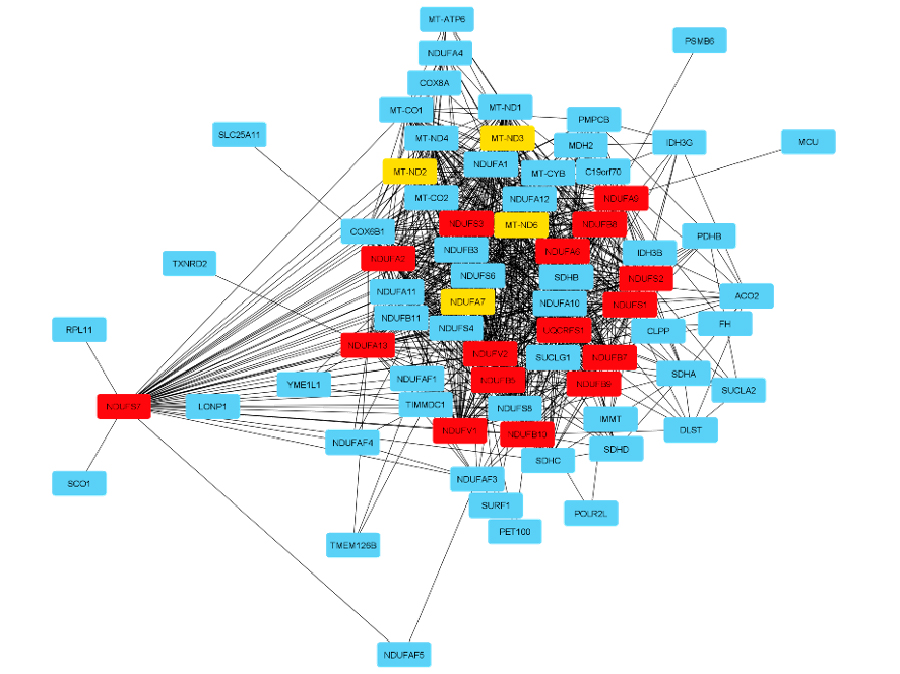
The top hub genes and the related neighbors are shown in Figure 2. Most of them are part of the NADH Ubiquinone Oxidoreductase family, like NDUFB7, NDUFS7, NDUFB8, NDUFA13, NDUFS2, NDUFV1, NDUFV2, NDUFS3, NDUFA9, NDUFA2, NDUFB5, NDUFA6, NDUFB9, NDUFB10, NDUFS1, and NDUFA7. In addition, we also identified as hub genes the Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase genes, like MTND6, MTND2, and MTND3. Another hub gene identified in our analysis is the Ubiquinol-Cytochrome C Reductase, Rieske Iron-Sulfur Polypeptide - UQCRFS1 gene.
Hub gene network and the related expanded subnetwork. The MCC method ranks the genes with a red-yellow scale, when red genes are the most relevant in the top 20 genes in the network.
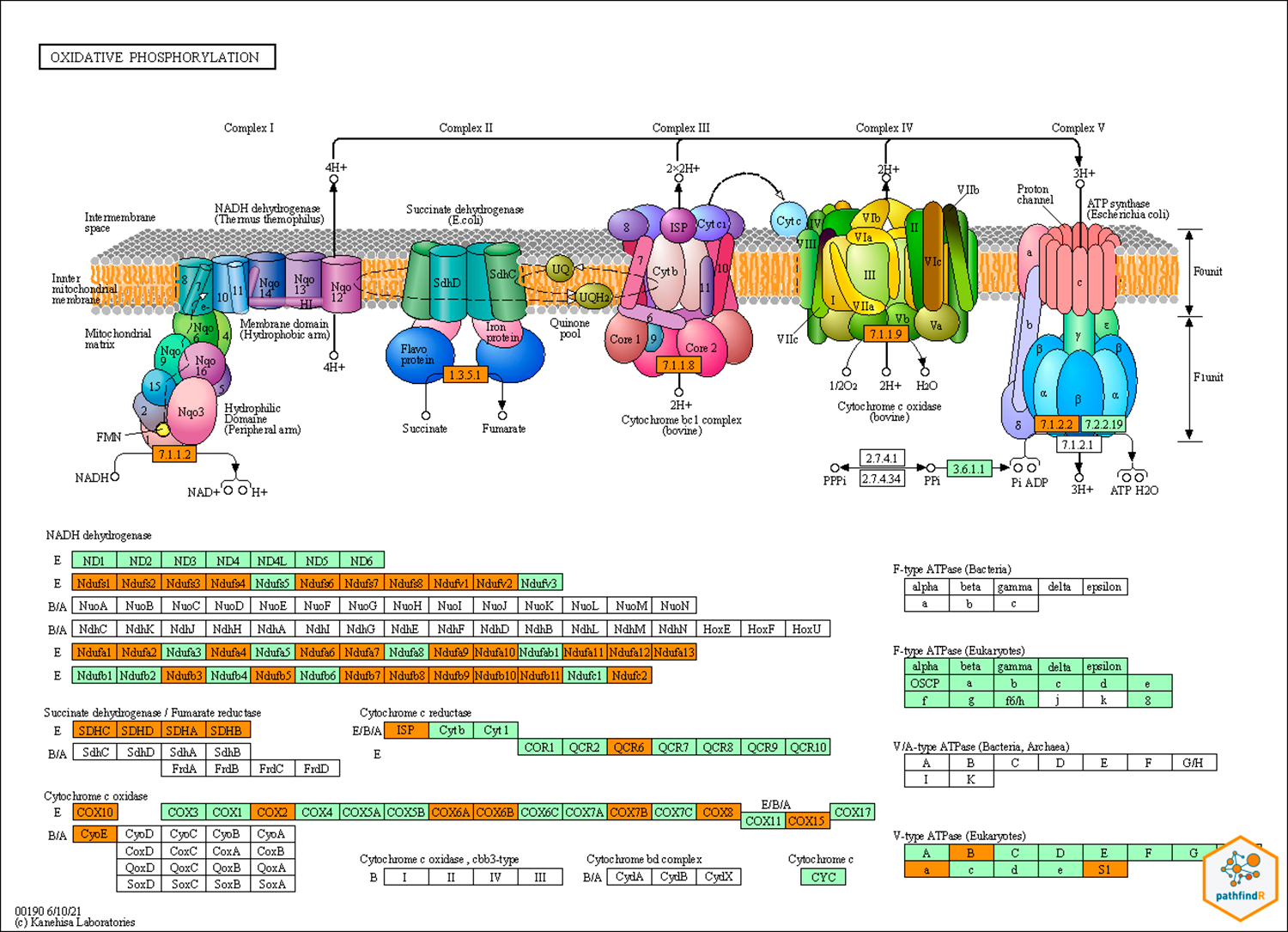
Regarding the pathway analysis, the most frequent genes were MAPK1, PIK3CA, PIK3R1, AKT1, KRAS, MAP2K1, NRAS, PRKCB, RAF1, and NFKB1. There were 114 enriched pathways related to the hearing loss gene list (Table 2). The top KEGG-related pathways are shown in Figure 3. We also constructed the KEGG maps to understand how the gene hub list affects the enriched pathways (Figure 4, Supplementar File 1 Supplementar File 1 - KEGG maps of the top enriched pathways. ).
KEGG map of the oxidative phosphorylation pathway. Green genes are the most important genes in the pathway. In orange, the genes that appear in our network.
Gene expression analysis showed NDUF genes (the top hub genes) to be up-regulated in MPS IIIB, while NDFUS7 is down-regulated in MPS I. NDUFV2 and NDUFS3 are not identified as differentially expressed. The same pattern is seen in the gene pathways list (Table 3).
Discussion
Several pathways involved in cell adhesion, proliferation and differentiation were enriched in our analysis. The Wnt signaling is the most enriched pathway, as it controls cellular events related to the formation of sensory hair cells during development [1515. Żak M, van Oort T, Hendriksen FG, Garcia M-I, Vassart G, Grolman W. LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea. Front Cell Neurosci. 2016;10:186. doi:10.3389/fncel.2016.00186.
https://doi.org/10.3389/fncel.2016.00186...
], and in cochlear formation and hair cell differentiation and polarization [1616. Waqas M, Zhang S, He Z, Tang M, Chai R. Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea. Front Med. 2016;10(3):237-249. doi:10.1007/s11684-016-0464-9.
https://doi.org/10.1007/s11684-016-0464-...
-1717. Warrick PD, Wardrop P, Sim DW. Sensorineural hearing loss in MELAS syndrome. J Laryngol Otol. 1997;111(3):279-281. doi:10.1017/s0022215100137089.
https://doi.org/10.1017/s002221510013708...
]. Other pathways, like PI3K/Akt, MAPK/ERK and EGFR and ERBB signaling were also enriched. Given that these pathways are related to formation, maintenance and regeneration activity of specialized cells, it is not surprising that these pathways are deranged in progressive degenerative diseases, as MPS.
Another set of differentially expressed genes in our analysis were related to mitochondrial function. This organelle has a role in oxidative phosphorylation, oxidative stress control, and apoptosis. The relationship between hearing loss and mitochondrial diseases has been discussed previously in the literature [1818. Hsu C-H, Kwon H, Perng C-L, Bai R-K, Dai P, Wong L-JC. Hearing loss in mitochondrial disorders. Ann N Y Acad Sci. 2005;1042:36-47. doi:10.1196/annals.1338.004.
https://doi.org/10.1196/annals.1338.004...
-2020. Elias TGA, Monsato RC, Amaral JB, Oyama LM, Maza PK, Penido NO. Evaluation of oxidative-stress pathway and recovery of sudden sensorineural hearing loss. Int Arch Otorhinolaryngol. 2021;25(3):e428-e432. doi:10.1055/s-0040-1714130.
https://doi.org/10.1055/s-0040-1714130...
]. Zwirner and Wilichowski demonstrated that there is a high incidence of 42% of sensorineural hearing loss in childrens with mitochondrial encephalomyopathies [2121. Zwirner P, Wilichowski E. Progressive sensorineural hearing loss in children with mitochondrial encephalomyopathies. Laryngoscope. 2001;111(3):515-21. doi:10.1097/00005537-200103000-00024.
https://doi.org/10.1097/00005537-2001030...
]. Besides, it was shown that causative mitochondrial DNA mutations appear in 5-10% of patients with post-lingual nonsyndromic hearing loss [2222. Jacobs HT, Hutchin TP, Käppi T, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13(1):26-33. doi:10.1038/sj.ejhg.5201250.
https://doi.org/10.1038/sj.ejhg.5201250...
].
Mitochondrial defects in MPS were also described in the literature. Martins and collaborators observed structurally abnormal mitochondria and impaired mitochondrial energy metabolism in a 5-month-old mouse model of MPS III C [2323. Martins C, Hůlková H, Dridi L, et al. Neuroinflammation, mitochondrial defects and neurodegeneration in mucopolysaccharidosis III type C mouse model. Brain. 2015;138(2):336-355. doi:10.1093/brain/awu355.
https://doi.org/10.1093/brain/awu355...
]. In another study, light microscopy of brain sections of 6-months-old mice with MPS III B showed the accumulation of mitochondrial ATP synthase subunit c in the brain [2424. Ryazantsev S, Yu W-H, Zhao H-Z, Neufeld EF, Ohmi K. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol Genet Metab. 2007;90(4):393-401. doi:10.1016/j.ymgme.2006.11.006.
https://doi.org/10.1016/j.ymgme.2006.11....
]. Alterations in mitochondria and lysosomes lead to neurological dysfunction and oxidative stress [2525. Fivenson EM, Lautrup S, Sun N, et al. Mitophagy in neurodegeneration and aging. Neurochem Int. 2017;109:202-209. doi:10.1016/j.neuint.2017.02.007.
https://doi.org/10.1016/j.neuint.2017.02...
], observed in some MPS types. Interestingly, Baixauli et al. [2626. Baixauli F, Acín-Pérez R, Villarroya-Beltrí C, et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 2015;22(3):485-498. doi:10.1016/j.cmet.2015.07.020.
https://doi.org/10.1016/j.cmet.2015.07.0...
] showed that mitochondrial deficiency impairs lysosome function, and disrupts endolysosomal trafficking pathways and autophagy, thus linking a primary mitochondrial dysfunction to a lysosomal disturbance. Mitochondrial dysfunction is emerging as a significant contributor to the pathophysiology of lysosomal storage disorders, like MPS [2727. Stepien KM, Roncaroli F, Turton N, et al. Mechanisms of mitochondrial dysfunction in lysosomal storage disorders: a review. J Clin Med. 2020;9(8):2596. doi:10.3390/jcm9082596.
https://doi.org/10.3390/jcm9082596...
].
The NADH:ubiquinone oxidoreductase subunits gene family appeared several times in our hub analysis. The NADH:ubiquinone oxidoreductase (complex I) is part of the respiratory complex and is a major source of reactive oxygen species (ROS) and an essential contributor to cellular oxidative stress. Moreover, ROS production has a relationship with several apoptotic and necrotic cell death pathways in auditory tissues [2828. de Beeck KO, Schacht J, Camp GV. Apoptosis in acquired and genetic hearing impairment: the programmed death of the hair cell. Hear Res. 2011;281(1-2):18-27. doi:10.1016/j.heares.2011.07.002.
https://doi.org/10.1016/j.heares.2011.07...
]. The subsequent apoptosis induction and elevated ROS formation are involved in developing several hearing loss impairments. In parallel, the involvement of ROS in MPS IVA [2929. Tsutsumi T, Nishida H, Noguchi Y, Komatzuzaki A, Kitamura K. Audiological findings in patients with myoclonic epilepsy associated with ragged-red fibres. J Laryngol Otol. 2001;115(10):777-781. doi:10.1258/0022215011909224.
https://doi.org/10.1258/0022215011909224...
-3030. Donida B, Marchetti DP, Biancini GB, et al. Oxidative stress and inflammation in mucopolysaccharidosis type IVA patients treated with enzyme replacement therapy. Biochim Biophys Acta. 2015;1852(5):1012-1019. doi:10.1016/j.bbadis.2015.02.004.
https://doi.org/10.1016/j.bbadis.2015.02...
] and MPS IIIB [3131. Donida B, Marchetti DP, Jacques CED, et al. Oxidative profile exhibited by Mucopolysaccharidosis type IVA patients at diagnosis: Increased keratan urinary levels. Mol Genet Metab Rep. 2017;11:46-53. doi:10.1016/j.ymgmr.2017.04.005.
https://doi.org/10.1016/j.ymgmr.2017.04....
] has been shown, but the role of the NDUF family in any type of MPS is not yet demonstrated.
Several genes related to complex I are up-regulated in MPS III B and down-regulated in MPS I - both of which present hearing loss in a significant portion of patients [3232. Villani GRD, Gargiulo N, Faraonio R, Castaldo S, Gonzalez Y Reyero E, Di Natale P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J Neurosci Res. 2007;85(3):612-622. doi:10.1002/jnr.21134.
https://doi.org/10.1002/jnr.21134...
-3636. Kiely BT, Kohler JL, Coletti HY, Poe MD, Escolar ML. Early disease progression of Hurler syndrome. Orphanet J Rare Dis. 2017;12(1):32. doi:10.1186/s13023-017-0583-7.
https://doi.org/10.1186/s13023-017-0583-...
]. In MPS III, previous studies demonstrated rates of hearing loss of 87% in MPS III A, 100% in MPS III B, 75% in MPS III C, and 25% in MPS IIID [3737. Zafeiriou DI, Savvopoulo-Augoustidou PA, Sewell A, et al. Serial magnetic resonance imaging findings in mucopolysaccharidosis IIIB (Sanfilippo’s syndrome B). Brain Dev. 2001;23(6):385-389. doi:10.1016/s0387-7604(01)00242-x.
https://doi.org/10.1016/s0387-7604(01)00...
-4040. Jansen ACM, Cao H, Kaplan P, et al. Sanfilippo syndrome type D: natural history and identification of 3 novel mutations in the GNS gene. Arch Neurol. 2007;64(11):1629-1634. doi:10.1001/archneur.64.11.1629.
https://doi.org/10.1001/archneur.64.11.1...
]. MPS III B’s higher proportion can be related to NDUF expression results, although more studies are needed to understand and validate this relationship.
Other genes found in our network analysis are related to succinate dehydrogenase, fumarate reductase, cytochrome c oxidase and reductase, and V-ATPase, which are all involved in the oxidative phosphorylation (Figure 4). A3, one of the four isoforms of subunit A V-ATPase, is required for secretory lysosome trafficking to the plasma membrane. It is also necessary to maintain the ionic concentration and pH of the endolymph that bathes the mechanosensory hair cells of the Corti organ in the inner ear [4141. Couloigner V, Teixeira M, Hulin P, et al. Effect of locally applied drugs on the pH of luminal fluid in the endolymphatic sac of guinea pig. Am J Physiol Regul Integr Comp Physiol. 2000;279(50):R1695-R1700. doi:10.1152/ajpregu.2000.279.5.R1695.
https://doi.org/10.1152/ajpregu.2000.279...
-4444. Stanković KM, Brown D, Alper SL, Adams JC. Localization of pH regulating proteins H+ATPase and exchanger in the guinea pig inner ear. Hear Res. 1997;114(1-2):21-34. doi:10.1016/s0378-5955(97)00072-5.
https://doi.org/10.1016/s0378-5955(97)00...
]. The V-ATPase is also present in the cochlea, and interdental cells are especially V-ATPase-rich [4444. Stanković KM, Brown D, Alper SL, Adams JC. Localization of pH regulating proteins H+ATPase and exchanger in the guinea pig inner ear. Hear Res. 1997;114(1-2):21-34. doi:10.1016/s0378-5955(97)00072-5.
https://doi.org/10.1016/s0378-5955(97)00...
]. Besides that, mutations in the subunit A4 or B1 are associated with sensorineural hearing loss [4343. Karet FE, Finberg KE, Nelson RD, et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21(1):84-90. doi:10.1038/5022.
https://doi.org/10.1038/5022...
]. Moreover, Santra and Amack, 2021 [4545. Santra P, Amack JD. Loss of vacuolar-type H+-ATPase induces caspase-independent necrosis-like death of hair cells in zebrafish neuromasts. Dis Model Mech. 2021;14(7):dmm048997. doi:10.1242/dmm.048997.
https://doi.org/10.1242/dmm.048997...
] have shown a specific role for V-ATPase inducing caspase-independent necrosis-like cell death in mechanosensory hair cells in neuromasts. Patients with mutations in specific V-ATPase subunits can develop sensorineural deafness. The mechanism involves modulation of the mitochondrial permeability transition pore, which regulates mitochondrial membrane potential, thus improving hair cell survival.
The majority of young MPS patients present mixed hearing loss (32%) and 16% sensorineural [4646. Vargas-Gamarra MF, de Paula-Vernetta C, Miñana IV, Ibañez-Alcañiz I, Cavallé-Garrido L, Alamar-Velazquez A. Audiological findings in children with mucopolysaccharidoses type i-iv. Acta Otorrinolaringol Esp (Engl Ed). 2017;68(5):262-268. doi:10.1016/j.otorri.2016.11.004.
https://doi.org/10.1016/j.otorri.2016.11...
]. Many studies exploring the mechanisms of hearing loss in MPS suggest a combination of conductive and sensorineural processes [33. Wolfberg J, Chintalapati K, Tomatsu S, Nagao K. Hearing loss in mucopolysaccharidoses: current knowledge and future directions. Diagnostics (Basel). 2020;10(8):554. doi:10.3390/diagnostics10080554.
https://doi.org/10.3390/diagnostics10080...
]. In this model, GAG accumulation leads to copious secretion and recurrent ear infection that, in conjunction with bone and cartilage deformities, contribute to conductive hearing loss. In addition, sensorineural hearing loss is caused by the death of hair cells. Our study sheds light into this second mechanism, suggesting a role for mitochondrial and V-ATPase dysfunction in the loss of hair cells [4747. Eaton AF, Merkulova M, Brown D. The H+-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am J Physiol Cell Physiol. 2021;320(3):C392-C414. doi:10.1152/ajpcell.00442.2020.
https://doi.org/10.1152/ajpcell.00442.20...
]. Even though we analyzed data from in vitro neural stem cells and not from the inner ear, one can suppose that the same mechanisms that lead to lysosomal storage-derived disturbance of mitochondrial function and V-ATPases in the brain are also present in other cells, such as cochlear hair cells, but in that case with specific consequences. Therefore, experimental studies in MPS animal models could test this hypothesis directly in the involved cells.
Conclusions
We identified several genes and biological pathways involved in ear development and hearing loss. These genes and biological pathways may serve as potential candidates for clinical and experimental studies to better understand hearing impairment mechanisms in lysosomal storage diseases, like MPS
References
- 1. Broek BTA, Smit AL, Boelens JJ, Hasselt PM. Hearing loss in patients with mucopolysaccharidoses ‐1 and ‐6 after hematopoietic cell transplantation: a longitudinal analysis. J Inherit Metab Dis 2020;43(6):1279-1287. doi:10.1002/jimd.12277.
» https://doi.org/10.1002/jimd.12277 - 2. Nagao K, Morlet T, Haley E, et al. Neurophysiology of hearing in patients with mucopolysaccharidosis type IV. Mol Genet Metab 2018;123(4):472-478. doi:10.1016/j.ymgme.2018.02.002.
» https://doi.org/10.1016/j.ymgme.2018.02.002 - 3. Wolfberg J, Chintalapati K, Tomatsu S, Nagao K. Hearing loss in mucopolysaccharidoses: current knowledge and future directions. Diagnostics (Basel) 2020;10(8):554. doi:10.3390/diagnostics10080554.
» https://doi.org/10.3390/diagnostics10080554 - 4. Isaacson JE, Vora NM. Differential diagnosis and treatment of hearing loss. Am Fam Physician 2003;68(6):1125-1132.
- 5. Köhler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res 2021;49(D1):D1207-D1217. doi:10.1093/nar/gkaa1043.
» https://doi.org/10.1093/nar/gkaa1043 - 6. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48(D1):D845-D855. doi:10.1093/nar/gkz1021.
» https://doi.org/10.1093/nar/gkz1021 - 7. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607-D613. doi:10.1093/nar/gky1131.
» https://doi.org/10.1093/nar/gky1131 - 8. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-2504. doi:10.1101/gr.1239303.
» https://doi.org/10.1101/gr.1239303 - 9. Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8(4 Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11.
» https://doi.org/10.1186/1752-0509-8-S4-S11 - 10. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21(16):3448-3449. doi:10.1093/bioinformatics/bti551.
» https://doi.org/10.1093/bioinformatics/bti551 - 11. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353-D361. doi:10.1093/nar/gkw1092.
» https://doi.org/10.1093/nar/gkw1092 - 12. Ulgen E, Ozisik O, Sezerman OU. pathfindR: An R Package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet 2019;10:858. doi:10.3389/fgene.2019.00858.
» https://doi.org/10.3389/fgene.2019.00858 - 13. R Core Team. R: A Language and Environment for Statistical Computing [Computer Software]. Vienna, Austria: R Foundation for Statistical Computing; 2017. https://www.R-project.org/
» https://www.R-project.org/ - 14. Soares LDF, Silva GCV, Kubaski F, Giugliani R, Matte U. MPSBase: Comprehensive repository of differentially expressed genes for mucopolysaccharidoses. Mol Genet Metab 2021;133(4):372-377. doi:10.1016/j.ymgme.2021.06.004.
» https://doi.org/10.1016/j.ymgme.2021.06.004 - 15. Żak M, van Oort T, Hendriksen FG, Garcia M-I, Vassart G, Grolman W. LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea. Front Cell Neurosci 2016;10:186. doi:10.3389/fncel.2016.00186.
» https://doi.org/10.3389/fncel.2016.00186 - 16. Waqas M, Zhang S, He Z, Tang M, Chai R. Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea. Front Med 2016;10(3):237-249. doi:10.1007/s11684-016-0464-9.
» https://doi.org/10.1007/s11684-016-0464-9 - 17. Warrick PD, Wardrop P, Sim DW. Sensorineural hearing loss in MELAS syndrome. J Laryngol Otol 1997;111(3):279-281. doi:10.1017/s0022215100137089.
» https://doi.org/10.1017/s0022215100137089 - 18. Hsu C-H, Kwon H, Perng C-L, Bai R-K, Dai P, Wong L-JC. Hearing loss in mitochondrial disorders. Ann N Y Acad Sci 2005;1042:36-47. doi:10.1196/annals.1338.004.
» https://doi.org/10.1196/annals.1338.004 - 19. Chennupati SK, Levi J, Loftus P, Jornlin C, Morlet T, O'Reilly RC. Hearing loss in children with mitochondrial disorders. Int J Pediatr Otorhinolaryngol 2011;75(12):1519-1524. doi:10.1016/j.ijporl.2011.08.019.
» https://doi.org/10.1016/j.ijporl.2011.08.019 - 20. Elias TGA, Monsato RC, Amaral JB, Oyama LM, Maza PK, Penido NO. Evaluation of oxidative-stress pathway and recovery of sudden sensorineural hearing loss. Int Arch Otorhinolaryngol 2021;25(3):e428-e432. doi:10.1055/s-0040-1714130.
» https://doi.org/10.1055/s-0040-1714130 - 21. Zwirner P, Wilichowski E. Progressive sensorineural hearing loss in children with mitochondrial encephalomyopathies. Laryngoscope 2001;111(3):515-21. doi:10.1097/00005537-200103000-00024.
» https://doi.org/10.1097/00005537-200103000-00024 - 22. Jacobs HT, Hutchin TP, Käppi T, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet 2005;13(1):26-33. doi:10.1038/sj.ejhg.5201250.
» https://doi.org/10.1038/sj.ejhg.5201250 - 23. Martins C, Hůlková H, Dridi L, et al. Neuroinflammation, mitochondrial defects and neurodegeneration in mucopolysaccharidosis III type C mouse model. Brain 2015;138(2):336-355. doi:10.1093/brain/awu355.
» https://doi.org/10.1093/brain/awu355 - 24. Ryazantsev S, Yu W-H, Zhao H-Z, Neufeld EF, Ohmi K. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol Genet Metab 2007;90(4):393-401. doi:10.1016/j.ymgme.2006.11.006.
» https://doi.org/10.1016/j.ymgme.2006.11.006 - 25. Fivenson EM, Lautrup S, Sun N, et al. Mitophagy in neurodegeneration and aging. Neurochem Int 2017;109:202-209. doi:10.1016/j.neuint.2017.02.007.
» https://doi.org/10.1016/j.neuint.2017.02.007 - 26. Baixauli F, Acín-Pérez R, Villarroya-Beltrí C, et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab 2015;22(3):485-498. doi:10.1016/j.cmet.2015.07.020.
» https://doi.org/10.1016/j.cmet.2015.07.020 - 27. Stepien KM, Roncaroli F, Turton N, et al. Mechanisms of mitochondrial dysfunction in lysosomal storage disorders: a review. J Clin Med 2020;9(8):2596. doi:10.3390/jcm9082596.
» https://doi.org/10.3390/jcm9082596 - 28. de Beeck KO, Schacht J, Camp GV. Apoptosis in acquired and genetic hearing impairment: the programmed death of the hair cell. Hear Res 2011;281(1-2):18-27. doi:10.1016/j.heares.2011.07.002.
» https://doi.org/10.1016/j.heares.2011.07.002 - 29. Tsutsumi T, Nishida H, Noguchi Y, Komatzuzaki A, Kitamura K. Audiological findings in patients with myoclonic epilepsy associated with ragged-red fibres. J Laryngol Otol 2001;115(10):777-781. doi:10.1258/0022215011909224.
» https://doi.org/10.1258/0022215011909224 - 30. Donida B, Marchetti DP, Biancini GB, et al. Oxidative stress and inflammation in mucopolysaccharidosis type IVA patients treated with enzyme replacement therapy. Biochim Biophys Acta 2015;1852(5):1012-1019. doi:10.1016/j.bbadis.2015.02.004.
» https://doi.org/10.1016/j.bbadis.2015.02.004 - 31. Donida B, Marchetti DP, Jacques CED, et al. Oxidative profile exhibited by Mucopolysaccharidosis type IVA patients at diagnosis: Increased keratan urinary levels. Mol Genet Metab Rep 2017;11:46-53. doi:10.1016/j.ymgmr.2017.04.005.
» https://doi.org/10.1016/j.ymgmr.2017.04.005 - 32. Villani GRD, Gargiulo N, Faraonio R, Castaldo S, Gonzalez Y Reyero E, Di Natale P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J Neurosci Res 2007;85(3):612-622. doi:10.1002/jnr.21134.
» https://doi.org/10.1002/jnr.21134 - 33. Silveira MRM, Buriti AKL, Martins AM, Gil D, Azevedo MF. Audiometric evaluation in individuals with mucopolysaccharidosis. Clinics (São Paulo) 2018;73:e523. doi:10.6061/clinics/2018/e523.
» https://doi.org/10.6061/clinics/2018/e523 - 34. Aldenhoven M, Wynn RF, Orchard PJ, et al. Long-term outcome of Hurler syndrome patients after hematopoietic cell transplantation: an international multicenter study. Blood 2015;125(13):2164-2172. doi:10.1182/blood-2014-11-608075.
» https://doi.org/10.1182/blood-2014-11-608075 - 35. Dualibi APFF, Martins AM, Moreira GA, Azevedo MF, Fujita RR, Pignatari SSN. The impact of laronidase treatment in otolaryngological manifestations of patients with mucopolysaccharidosis. Braz J Otorhinolaryngol 2016;82(5):522-528. doi:10.1016/j.bjorl.2015.09.006.
» https://doi.org/10.1016/j.bjorl.2015.09.006 - 36. Kiely BT, Kohler JL, Coletti HY, Poe MD, Escolar ML. Early disease progression of Hurler syndrome. Orphanet J Rare Dis 2017;12(1):32. doi:10.1186/s13023-017-0583-7.
» https://doi.org/10.1186/s13023-017-0583-7 - 37. Zafeiriou DI, Savvopoulo-Augoustidou PA, Sewell A, et al. Serial magnetic resonance imaging findings in mucopolysaccharidosis IIIB (Sanfilippo’s syndrome B). Brain Dev 2001;23(6):385-389. doi:10.1016/s0387-7604(01)00242-x.
» https://doi.org/10.1016/s0387-7604(01)00242-x - 38. Buhrman D, Thakkar K, Poe M, Escolar ML. Natural history of Sanfilippo syndrome type A. J Inherit Metab Dis 2014;37(30):431-437. doi:10.1007/s10545-013-9661-8.
» https://doi.org/10.1007/s10545-013-9661-8 - 39. Ruijter GJG, Valstar MJ, van de Kamp JM, et al. Clinical and genetic spectrum of Sanfilippo type C (MPS IIIC) disease in The Netherlands. Mol Genet Metab 2008;93(2):104-11. doi:10.1016/j.ymgme.2007.09.011.
» https://doi.org/10.1016/j.ymgme.2007.09.011 - 40. Jansen ACM, Cao H, Kaplan P, et al. Sanfilippo syndrome type D: natural history and identification of 3 novel mutations in the GNS gene. Arch Neurol 2007;64(11):1629-1634. doi:10.1001/archneur.64.11.1629.
» https://doi.org/10.1001/archneur.64.11.1629 - 41. Couloigner V, Teixeira M, Hulin P, et al. Effect of locally applied drugs on the pH of luminal fluid in the endolymphatic sac of guinea pig. Am J Physiol Regul Integr Comp Physiol 2000;279(50):R1695-R1700. doi:10.1152/ajpregu.2000.279.5.R1695.
» https://doi.org/10.1152/ajpregu.2000.279.5.R1695 - 42. Ferrary E, Sterkers O. Mechanisms of endolymph secretion. Kidney Int Suppl 1998;65:S98-S103.
- 43. Karet FE, Finberg KE, Nelson RD, et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 1999;21(1):84-90. doi:10.1038/5022.
» https://doi.org/10.1038/5022 - 44. Stanković KM, Brown D, Alper SL, Adams JC. Localization of pH regulating proteins H+ATPase and exchanger in the guinea pig inner ear. Hear Res 1997;114(1-2):21-34. doi:10.1016/s0378-5955(97)00072-5.
» https://doi.org/10.1016/s0378-5955(97)00072-5 - 45. Santra P, Amack JD. Loss of vacuolar-type H+-ATPase induces caspase-independent necrosis-like death of hair cells in zebrafish neuromasts. Dis Model Mech 2021;14(7):dmm048997. doi:10.1242/dmm.048997.
» https://doi.org/10.1242/dmm.048997 - 46. Vargas-Gamarra MF, de Paula-Vernetta C, Miñana IV, Ibañez-Alcañiz I, Cavallé-Garrido L, Alamar-Velazquez A. Audiological findings in children with mucopolysaccharidoses type i-iv. Acta Otorrinolaringol Esp (Engl Ed) 2017;68(5):262-268. doi:10.1016/j.otorri.2016.11.004.
» https://doi.org/10.1016/j.otorri.2016.11.004 - 47. Eaton AF, Merkulova M, Brown D. The H+-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am J Physiol Cell Physiol 2021;320(3):C392-C414. doi:10.1152/ajpcell.00442.2020.
» https://doi.org/10.1152/ajpcell.00442.2020
-
Funding
We would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the fellowships provided GCVS (Process no.: 148615/2018-0), PB, AG, and UM (Process no.: 312714/2018-1) . We also thank FIPE/HCPA (Project Number 2018-0594), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES for financial support.
Supplementary Material
The following online material is available for this article:
Supplementar File 1 - KEGG maps of the top enriched pathways.
Publication Dates
-
Publication in this collection
13 May 2022 -
Date of issue
2022
History
-
Received
31 Dec 2021 -
Accepted
30 Mar 2022