ABSTRACT:
This study evaluated the genetic diversity and antimicrobial susceptibility of Staphylococcus aureus isolates from dairy cows in Minas Gerais, Brazil. Thirty-seven isolates from five municipalities (8 herds) were genotyped using multi-locus sequence typing (MLST) and susceptibility to 12 antimicrobial agents was tested using the disk diffusion method. High resistance rates for penicillin [75.68% (28/37)], ampicillin [70.27% (26/37)], and tetracycline [70.27% (26/37)] were detected. Multidrug resistance was observed in seven [18.92% (7/37)] isolates, and two were suggestive of Methicillin-resistant Staphylococcus aureus (MRSA). Among the 37 isolates, 33 novel sequence types (ST) and two known STs (ST126 and ST746) were identified in MLST. The clonal complexes more frequently observed were: CC97 [78.38%; (29/37)], CC1 [8.11%; (3/37)] and CC5 [5.40%; (2/37)]. Minimum‐spanning tree (MST) analysis according to data from municipalities, herds, and resistance patterns for all isolates did not show any clustering pattern. However, the MST comparing all Brazilian S. aureus isolates deposited in the PubMLST database and from this study depicted an association between the genotype and strain origin (clinical sample). Isolates from this study that belong to CC97 were close to database isolates from milk and dairy products, while those that belong to CC1 and CC5 were close to database isolates from human sources and the environment of dairy farms or industries. In conclusion, our results showed a high rate of resistance to penicillins and tetracyclines and great genetic diversity among the S. aureus isolates from bovine mastitis genotyped in the present study.
Key words:
staphylococci; MLST; multidrug resistance; dairy industry; zoonosis
RESUMO:
Este estudo teve como objetivo avaliar a diversidade genética e a suscetibilidade a antimicrobianos de cepas de Staphylococcus aureus isoladas de vacas leiteiras em Minas Gerais, Brasil. Trinta e sete cepas provenientes de cinco municípios (oito rebanhos) foram genotipadas usando a técnica multi-locus sequence typing (MLST) e a suscetibilidade a 12 antimicrobianos foi avaliada pelo método de difusão em disco. Foram detectadas altas taxas de resistência para penicilina [75,68% (28/37)], ampicilina [70,27% (26/37)] e tetraciclina [70,27% (26/37)] entre os isolados. A multirresistência foi observada em sete [18,92% (7/37)] isolados e dois foram classificados como sugestivos de Staphylococcus aureus resistente à meticilina (MRSA). Entre os 37 isolados, 33 novos sequence types (ST) e dois STs conhecidos (ST126 e ST746) foram identificados pelo MLST. Os complexos clonais mais frequentemente observados foram: CC97 [78,38%; (29/37)], CC1 [8,11%; (3/37)] e CC5 [5,40%; (2/37)]. Foi construída uma minimum‐spanning tree (MSTs) com todos isolados estudados e esta não mostrou padrão de agrupamento quando comparada com dados epidemiológicos como municípios e rebanho, do qual foi isolado, e perfis de resistência a antimicrobianos. Uma segunda MST foi construída comparando os isolados deste estudo e todas as cepas de S. aureus depositadas no banco de dados PubMLST provenientes do Brasil, que mostrou associação entre o genótipo, STs e a origem da cepa. Foi possível observar entre os isolados do estudo que, aqueles que pertenciam ao CC97, eram geneticamente mais próximos das cepas depositadas no PubMLST isoladas de leite e de produtos lácteos, enquanto aqueles que pertenciam aos CC1 e CC5 estavam mais próximos a cepas isoladas de humanos ou do ambiente de fazendas e indústrias de laticínios. Em conclusão, nossos resultados mostraram um alto índice de resistência às penicilinas e tetraciclinas e grande diversidade genética entre as cepas de S. aureus isoladas de casos de mastite bovina.
Palavras-chave:
estafilococos; MLST; multirresistência; zoonose; setor lácteo
INTRODUCTION:
Staphylococcus aureus is a pathogen of humans and animals and is considered an important public and animal health issue (AIRES-DE-SOUSA et al., 2007AIRES-DE-SOUSA, M. et al. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Applied and Environmental Microbiology, 2007. v.73, n.12, p.3845-3849. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1932710/ >. Accessed: Jan. 15, 2021. doi: 10.1128/AEM.00019-07.
https://www.ncbi.nlm.nih.gov/pmc/article...
; PÉREZ et al., 2020PÉREZ, V. K. C. et al. Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. Journal of Global Antimicrobial Resistance, 2020. v.22, p.792-802. Available from: <Available from: https://doi.org/10.1016/j.jgar.2020.06.010 >. Accessed: Jan. 17, 2021. doi: 10.1016/j.jgar.2020.06.010.
https://doi.org/10.1016/j.jgar.2020.06.0...
; RAINARD et al., 2017RAINARD, P. et al. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transboundary and Emerging Diseases, 2017. v.65, n.1, p.149-165. Available from: <Available from: https://onlinelibrary.wiley.com/doi/10.1111/tbed.12698 >. Accessed: Jan. 17, 2021. doi: 10.1111/tbed.12698.
https://onlinelibrary.wiley.com/doi/10.1...
; SOBRAL et al., 2012SOBRAL, D. et al. High throughput multiple locus variable number of tandem repeat analysis (MLVA) of Staphylococcus aureus from human, animal and food sources. PLoS ONE, 2012. v.7, n.5, p.e33967. Available from: <Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033967 >. Accessed: Jan. 17, 2021. doi: 10.1371/journal.pone.0033967.
https://journals.plos.org/plosone/articl...
). Due to its high capacity of adaptation to the host and genetic diversity, specific lineages evolved to infect particular mammalian species; although, transmission between species, including zoonotic transmission, has been reported (SUNG et al., 2008SUNG, J. M. L.; LLOYD, D. H.; LINDSAY, J. A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology, 2008. v.154, n.7, p.1949-1959. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.2007/015289-0#tab2 >. Accessed: Jan. 22, 2021. doi: 10.1099/mic.0.2007/015289-0.
https://www.microbiologyresearch.org/con...
). The zoonotic potential of S. aureus is intensified taking into account its great ability to acquire resistance to several antimicrobials, with special attention to methicillin-resistant Staphylococcus aureus (MRSA). MRSA is historically associated with hospitals and other health services; however, recently, these isolates have also emerged as a widespread cause of infections in the community and animals (KINROSS et al., 2017KINROSS, P. et al. Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Surveillance and Outbreak Report, 2017. v.22, n.44, p.1-13. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5710135/ >. Accessed: Jan. 17, 2021. doi: 10.2807/1560-7917.ES.2017.22.44.16-00696.
https://www.ncbi.nlm.nih.gov/pmc/article...
).
In animal health, S. aureus stands out as one of the most important pathogens of dairy cattle (VIGUIER et al., 2009VIGUIER, C. et al. Mastitis detection: current trends and future perspectives. Trends in Biotechnology, 2009. v.27, n.8, p.486-493. Available from: <Available from: https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(09)00112-7 >. Accessed: Jan. 14, 2021. doi: 10.1016/j.tibtech.2009.05.004.
https://www.cell.com/trends/biotechnolog...
; VLIEGHER, DE et al., 2012VLIEGHER, S. DE et al. Invited review: Mastitis in dairy heifers: Nature of the disease, potential impact, prevention, and control. Journal of Dairy Science, 2012. v.95, n.3, p.1025-1040. Available from: <Available from: http://dx.doi.org/10.3168/jds.2010-4074 >. Accessed: Jan. 11, 2021. doi: 10.3168/jds.2010-4074.
http://dx.doi.org/10.3168/jds.2010-4074...
), responsible for causing bovine mastitis. S. aureus mastitis causes milk production losses of 2.3 kg/day, in addition to costs with discarded milk, diagnosis, and treatment, future milk production loss, premature culling, and replacement of cows (HEIKKILÄ et al., 2018HEIKKILÄ, A. M. et al. Pathogen-specific production losses in bovine mastitis. Journal of Dairy Science, 2018. v.101, n.10, p.9493-9504. Available from: <Available from: http://dx.doi.org/10.3168/jds.2018-14824 >. Accessed: Jan. 14, 2021. doi: 10.3168/jds.2018-14824.
http://dx.doi.org/10.3168/jds.2018-14824...
). In Brazil, the agent is highly prevalent in dairy cattle herds, and in the state of Minas Gerais, the prevalence in herds ranging from 28 to 93% have been reported (CUNHA, et al., 2016CUNHA, A. F. DA et al. Prevalence, etiology and risk factors of clinical mastitis in dairy cattle of Viçosa-MG. Acta Veterinaria Brasilica, 2016. v.10, n.1, p.48-54.; MESQUITA et al., 2019MESQUITA, A. A. et al. Staphylococcus aureus and streptococcus agalactiae: Prevalence, resistance to antimicrobials, and their relationship with the milk quality of dairy cattle herds in Minas Gerais state, Brazil. Pesquisa Veterinaria Brasileira, 2019. v.39, n.5, p.308-316. Available from: <Available from: https://www.scielo.br/j/pvb/a/4gysmHDfd3wqNh3YNtJnYBx/?lang=en >. Accessed: Jan. 16, 2021. doi: 10.1590/1678-5150-PVB-5821.
https://www.scielo.br/j/pvb/a/4gysmHDfd3...
).
S. aureus is mainly associated with subclinical mastitis cases, causing high somatic cell counts (CCS) detection in milk (RAINARD et al., 2017RAINARD, P. et al. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transboundary and Emerging Diseases, 2017. v.65, n.1, p.149-165. Available from: <Available from: https://onlinelibrary.wiley.com/doi/10.1111/tbed.12698 >. Accessed: Jan. 17, 2021. doi: 10.1111/tbed.12698.
https://onlinelibrary.wiley.com/doi/10.1...
). The infection among animals is primarily transmitted during the milking process (KULKARNI; KALIWAL, 2013KULKARNI, A. G.; KALIWAL, B. B. Bovine mastitis: A review. International Journal of Recent Scientific Research, 2013. v.4, n.5, p.543-548. Available from: <Available from: http://www.recentscientific.com/bovine-mastitis-review >. Accessed: Jan. 11, 2021.
http://www.recentscientific.com/bovine-m...
; RAINARD et al., 2017), and the bacteria then spread furtively within the herd. To understand the dynamic of the disease transmission, reservoirs of infections and to propose more effective control measures, it is critical to perform classical and molecular epidemiological studies, which allow the assessment of the frequency, distribution, and risk factors associated with staphylococcal mastitis, as well as the characterization of the isolates involved (TIBAYRENC, 2009TIBAYRENC, M. Microbial molecular epidemiology : An overview. In: CAUGANT, D. A. (Org.). Molecular Epidemiology of Microorganisms. 1. ed. Totowa, New Jersey: Humana Press, 2009, p. 1-12.). Furthermore, information on the susceptibility profile of S. aureus is also critical for decision making regarding mastitis therapies, health surveillance actions and the evaluation of antimicrobial use in animal production (PÉREZ et al., 2020PÉREZ, V. K. C. et al. Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. Journal of Global Antimicrobial Resistance, 2020. v.22, p.792-802. Available from: <Available from: https://doi.org/10.1016/j.jgar.2020.06.010 >. Accessed: Jan. 17, 2021. doi: 10.1016/j.jgar.2020.06.010.
https://doi.org/10.1016/j.jgar.2020.06.0...
). Molecular epidemiology studies along antimicrobial resistance evaluation in S. aureus from animal origin can drive the promotion of actions focused on the One Health approach, besides the evaluation of the interface between specific-human and specific-animal isolates.
One of the most used epidemiological molecular techniques for S. aureus is the multi-locus sequence typing (MLST), which is a sequencing-based genotyping method that assesses the polymorphisms in seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL), providing unique allelic profiles known as sequence types (STs). The level of discrimination (resolution) of MLST allows the assessment of a detailed picture of the global dissemination of this pathogen, supporting insights into its origin, pathogenicity, and evolution (SAUNDERS; HOLMES, 2007SAUNDERS, N. A.; HOLMES, A. Multilocus sequence typing (MLST) of Staphylococcus aureus. Methods in Molecular Biology, 2007. v.391, p.71-85. Available from: <Available from: https://link.springer.com/protocol/10.1007%2F978-1-59745-468-1_6 >. Accessed: Jan. 18, 2021. doi: 10.1007/978-1-59745-468-1_6.
https://link.springer.com/protocol/10.10...
). Thus, the present study aimed (i) to evaluate the genetic diversity of S. aureus isolated from dairy cows from Minas Gerais state, Brazil, using MLST, (ii) to determine the antimicrobial susceptibility profiles of these isolates, and (iii) the possible association between these variables and epidemiological data of the isolates.
MATERIALS AND METHODS:
Isolates
Thirty-seven (37) isolates of S. aureus previously isolated from milk samples of dairy cows with mastitis and S. aureus ATCC 25923T were used in the present study. Isolation and microbiological characterization of the isolates were performed according to described by BRITO & BRITO (1999BRITO, M. A. V. P. E; BRITO, J. R. F. Diagnóstico Microbiológico da Mastite. Juiz de Fora, MG: Embrapa Gado de Leite, 1999.).
The isolates belong to the Collection of Microorganisms of Interest to Agroindustry and Livestock (Embrapa Gado de Leite, Brazil), and were isolated between 2009 and 2011 from eight herds localized in five municipalities of Minas Gerais state. The municipalities were: Bias Fortes (Herd A, n=3 isolates); Bicas (Herd H, n=4); Lima Duarte (Herd D, n=10), Rio Preto (Herd E, n=5; Herd F, n=8; Herd G, n=2), and Santa Rita do Ibitipoca, (Herd B, n=3; Herd C, n=2). The geographical distribution of the municipalities and the distance, in kilometers (Km), between them are shown in figure 1.
Geographical distribution of the municipalities where Staphylococcus aureus isolates were collected from cows with mastitis, in Minas Gerais, Brazil, 2009-2011. The values on the lines refer to the distance in kilometers (Km) by official roads, shown by Google Maps, and not by the distance in a straight line.
Antimicrobial susceptibility test
Twelve antimicrobial agents were used to assess the antimicrobial susceptibility of the isolates using the disk diffusion method, according to the VET01‐A4 manual of the Clinical and Laboratory Standards Institute (CLSI, 2018CLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 5th ed. CLSI standard VET01. 5th. ed. Wayne: Clinical and Laboratory Standards Institute, 2018.). Antimicrobial agents tested and disk concentrations are described in table 1. To classify the isolates as resistant, intermediate, or sensitive to the tested antimicrobials, the CLSI manual VET08 was used (CLSI, 2018bCLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals VET08. 4th. ed. Wayne: Clinical and Laboratory Standards Institute , 2018b.). S. aureus ATCC 25923T was used for quality control in the susceptibility testing.
Multidrug resistance (MDR) was defined as resistance to three or more antimicrobial groups (MAGIORAKOS et al., 2011MAGIORAKOS, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 2011. v.18, n.3, p.268-281. Available from: <Available from: http://dx.doi.org/10.1111/j.1469-0691.2011.03570.x >. Accessed: Jan. 15, 2021. doi: 10.1111/j.1469-0691.2011.03570.x.
http://dx.doi.org/10.1111/j.1469-0691.20...
). The antimicrobial groups were as follows: penicillins (ampicillin, oxacillin, and penicillin G); cephems (cephalothin and ceftiofur); lincosamides (clindamycin); macrolides (erythromycin); quinolones (enrofloxacin); aminoglycosides (gentamicin); folate pathway inhibitors (sulfonamide and trimethoprim/sulfamethoxazole); and tetracyclines (tetracycline).
MLST
MLST was performed based on the DNA sequences of seven conserved housekeeping genes, arcC (carbamate kinase), aroE (shikimate dehydrogenase), glpF (glycerol kinase), gmk (guanylate kinase), pta (phosphate acetyltransferase), tpi (triosephosphate isomerase), and yqiL (acetyl coenzyme A acetyltransferase), which were amplified using specific primers as described by Enright et al. (2000ENRIGHT, M. C. et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology, 2000. v.38, n.3, p.1008-1015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ >. Accessed: Jan. 12, 2021. doi: 10.1128/JCM.38.3.1008-1015.2000.
https://www.ncbi.nlm.nih.gov/pmc/article...
).
The DNA sequences were obtained with an ABI-3500 automatic sequencer (Applied Biosystems, Foster, Californian, USA). The quality of sequence was evaluated using Phred software (reliability index > 20) (EWING et al., 1998EWING, B. et al. Base-calling of automated sequencer races using phred. I. Accuracy assessment. Genome Research, 1998. v.8, n.3, p.175-85. Available from: <Available from: https://genome.cshlp.org/content/8/3/175.long >. Accessed: Jan. 16, 2021. doi: 10.1101/gr.8.3.175.
https://genome.cshlp.org/content/8/3/175...
) and the consensus sequences were determined using the program CAP3 (HUANG; MADAN, 1999HUANG, X.; MADAN, A. CAP3: A DNA sequence assembly program. Genome Research, 1999. v.9, n.9, p.868-877. Available from: < Available from: https://www.genome.orgwww.genome.org >. Accessed: Jan. 18, 2021. doi: 10.1101/gr.9.9.868.
https://www.genome.orgwww.genome.org...
).
Alleles and STs were determined by comparing the sequences obtained with those deposited in PubMLST online database (https://pubmlst.org/). Alleles sequences were aligned using MEGA-X version 10.1.8 (TAMURA, STECHER, KUMAR, 2020) to assess the polymorphisms observed.
Isolates that shared four or more identical alleles were grouped in the same clonal complex (CC), according to the PubMLST database (https://pubmlst.org/organisms/staphylococcus-aureus/clonal-complexes). The Hunter and Gaston Diversity Index (HGDI) (HUNTER; GASTON, 1988HUNTER, P. R.; GASTON, M. A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. Journal of Clinical Microbiology, 1988. v.26, n.11, p.2465-2466. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC266921/ >. Accessed: Jan. 11, 2021. doi: 10.1128/jcm.26.11.2465-2466.1988.
https://www.ncbi.nlm.nih.gov/pmc/article...
) was calculated for each locus and MLST (http://insilico.ehu.eus/mini_tools/discriminatory_power/index.php).
To evaluate population structure and patterns of evolution, genetic comparisons among the isolates were performed using the goeEburst algorithm (https://online.phyloviz.net/index) (FRANCISCO et al., 2009FRANCISCO, A. P. et al. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics, 2009. v.10, p.1-15. Available from: <Available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-10-152 >. Accessed: Jan. 20, 2021. doi: 10.1186/1471-2105-10-152.
https://bmcbioinformatics.biomedcentral....
). The same software was used to build a minimum‐spanning tree (MST) and to assess possible clustering patterns of the isolates, considering the antimicrobial susceptibility profiles, herds, MDR, and municipalities. Isolates were also compared with all 336 S. aureus isolates from Brazil deposited in the PubMLST database (access on 30th March 2021), considering STs, CCs and resistance to methicillin.
RESULTS:
Antimicrobial susceptibility
The percentages of isolates classified as resistant, intermediate, or susceptible for each antimicrobial tested are shown in table 1. Resistance was observed mainly to penicillin [75.68% (28/37)], ampicillin [70.27% (26/37)], and tetracycline [70.27% (26/37)].
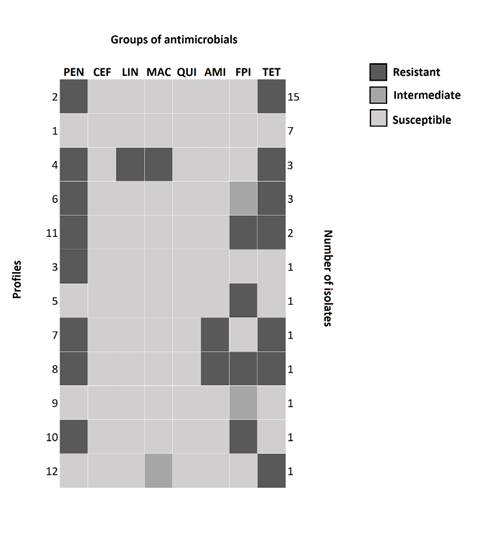
Eleven susceptibility profiles were constructed based on the antimicrobial groups (Figure 2). Multidrug resistance was observed in seven [18.92% (7/37)] isolates.
Antimicrobial susceptibility profiles of Staphylococcus aureus isolated from cows with mastitis, in Minas Gerais, Brazil, 2009-2011. Penicillins (PEN), Cephalosporins (CEF), Quinolones (QUI), Tetracyclines (TET), Macrolides (MAC), Lincosamides (LIN), and Folate Pathway Inhibitors (FPI). Enviar figura em .tiff com pelo menos 300 dpi.
MLST
Thirty-five (35) STs were identified among the 37 genotyped isolates, being two previously described (ST126; n=3; ST746; n=1) in the PubMLST database (pubmlst.org/saureus/) and 33 classified as novel STs. These novel STs were classified based on either the presence of a novel allele not described in the PubMLST database [32/33, (96.97%)] or a unique combination of known alleles [(1/33, 3.03%)]. The novel alleles were characterized mostly by nonsynonymous point mutations. The number of alleles per locus or STs, the alleles or ST most frequents, and the HGDI values are shown in table 2. The number of alleles per locus varied from 7 (pta) to 22 (yqiL) and the number of novel alleles per locus from 4 (pta) to 15 (yqiL).
Three clonal complexes were observed: CC97 [78.38%; (29/37)], CC1 [8.11%; (3/37)], CC5 [5.40%; (2/37)]; whereas three (5.40%) isolates could not be classified in a clonal complex. Different STs and CCs were found in the same herd (Table 3).
The isolates from this study were compared with each other (Figure 3a) and with all 336 S. aureus isolates from Brazil available in the MLST database (Figure 3b) using MSTs. These MLST data were obtained from Brazilian isolates obtained between 1997 and 2017 and distributed in all Brazilian regions: Southeast [233/336 (36.35%)], Midwest [61/336 (18.15%)], Northeast [19/336 (5.65%)], South [10/336 (2,98%)], and North [3/336 (0.89%)]. For ten (2.98%) isolates information on geographical origin was not available. These isolates were obtained from milk or dairy products (cow, goat, sheep, or buffalo) [101/336 (30.06%)], the environment of dairy farms or dairy industries [23/336 (6.85%)], animals [3/336 (0.89%)], other sources (mainly human disease cases) [24/336 (24.40%)] and unknowing sources [127/336 (37.80%)]. Among the isolates, it was observed 115 different STs.
(a) Minimum-spanning trees (MST) of the 37 isolates of Staphylococcus aureus isolated from cows with mastitis in dairy herds in Minas Gerais state, 2009-2011, and compared with epidemiological data of municipalities and herds. (b) MST generated with MLST data of all Brazilian entries of Staphylococcus aureus available in PubMLST (https://pubmlst.org/) and isolates of this study, associated with isolates sources. Circles represent clonal complexes reported in this study. Only isolates from this study are identified by numbers. Both MSTs presented were performed using the goeBURST algorithm available online (https://online.phyloviz.net/index).
MST analysis according to the municipality, herd, and antimicrobial resistance profiles considering only the isolates from this study did not show any clustering pattern. Conversely, MST performed using the isolates from this study together with the other Brazilian S. aureus isolates deposited in the PubMLST showed a clustering pattern for the source of isolation. Most of the S. aureus from this study were close to Brazilian isolates previously isolated from milk and dairy products (CC97); although, some isolates exhibited ST similar to isolates from other sources, such as human staphylococcal diseases (CC5) or dairy farms and industries environment (CC1).
DISCUSSION:
Genetic characterization and antimicrobial resistance assessment of pathogens associated with bovine mastitis are fundamental to understanding the epidemiology of the disease, routes of transmission, reservoirs, zoonotic potential, and trace control measures. For this, it is necessary to use typing techniques with high discriminatory power and good epidemiological concordance (TIBAYRENC, 2009TIBAYRENC, M. Microbial molecular epidemiology : An overview. In: CAUGANT, D. A. (Org.). Molecular Epidemiology of Microorganisms. 1. ed. Totowa, New Jersey: Humana Press, 2009, p. 1-12.), in addition to monitoring drug susceptibility. In this study, using the MLST technique, high genetic diversity was reported among S. aureus isolates from bovine mastitis, and of the 35 STs identified, 33 (94.28%) STs have not been previously described (Table 2).
Many of these new STs were characterized by point mutations in known alleles, which agrees with a study by Feil et al. (2003FEIL, E. J. et al. How clonaliIs Staphylococcus aureus? Journal of Bacteriology, 2003. v.185, n.11, p.3307-3316. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC155367/ >. Accessed: Jan. 17, 2021. doi: 10.1128/JB.185.11.3307-3316.2003.
https://www.ncbi.nlm.nih.gov/pmc/article...
), which concluded that point mutations give rise to new alleles at least 15-fold more frequently than does recombination. However, given that MLST is a technique based on sequencing housekeeping genes, this significant number of new alleles and STs observed in this study was not expected. Therefore, it is important to speculate on the reasons that may be accelerating the microevolution of this pathogen in Brazil, probably for better adaptation to the bovine mammary gland. A possible factor is the great spread of S. aureus in the Brazilian herds, considering the milk production profile and the intense animal trade between herds (RABELLO et al., 2007RABELLO, R. F. et al. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. Journal of Medical Microbiology, 2007. v.56, n.11, p.1505-1511. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.47357-0#tab2 >. Accessed: Jan. 14, 2021. doi: 10.1099/jmm.0.47357-0.
https://www.microbiologyresearch.org/con...
). Especially at Minas Gerais - the state of origin of the studied isolates - this intense trade, combined with inefficient mastitis control measures on farms, favors the constant transference of S. aureus among cows and herds, which may increase the selection pressure on the pathogen and, thereby, the emergence of new STs, as observed.
Conversely, it is also important to consider that the new STs reported in the present study may be quite common in Brazilian herds (mainly in Minas Gerais state), however, had not yet been described, since very few Brazilian isolates are available at PubMLST database. Indeed, there are only 336 (distributed in 115 different STs) Brazilian S. aureus isolates deposited in PubMLST of 35,737 S. aureus MLST total records. Moreover, of these 336 STs of Brazilian origin deposited, only 47 were from bovine mastitis isolates (24 STs). This highlighted the great scientific contribution of this study to understanding the genetic diversity and epidemiology of mastitis caused by S. aureus in Brazil.
The great number of new alleles and STs observed also resulted in a high genetic diversity among the typed isolates, which precluded the observation of clustering patterns that could indicated transmission routes or sources of infection among herds and municipalities. However, it was possible to observe that most of the isolates typed belonged to ST126 and ST746 or were very close to them (Figure 2b). These STs are poorly distributed lineages of S. aureus around the world and are associated with mastitis in ruminates (AIRES-DE-SOUSA et al., 2007AIRES-DE-SOUSA, M. et al. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Applied and Environmental Microbiology, 2007. v.73, n.12, p.3845-3849. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1932710/ >. Accessed: Jan. 15, 2021. doi: 10.1128/AEM.00019-07.
https://www.ncbi.nlm.nih.gov/pmc/article...
) but mainly described in studies realized in Brazil (AIRES-DE-SOUSA et al., 2007; RABELLO et al., 2007RABELLO, R. F. et al. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. Journal of Medical Microbiology, 2007. v.56, n.11, p.1505-1511. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.47357-0#tab2 >. Accessed: Jan. 14, 2021. doi: 10.1099/jmm.0.47357-0.
https://www.microbiologyresearch.org/con...
; SILVA et al., 2013SILVA, N. C. C. et al. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. Journal of Dairy Science, 2013. v.96, n.11, p.6856-6862. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/24054305/ >. Accessed: Jan. 17, 2021. doi: 10.3168/jds.2013-6719.
https://pubmed.ncbi.nlm.nih.gov/24054305...
) although ST126 isolates have been described as causing mastitis in the United States (SMITH et al., 2005SMITH, E. M. et al. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. Journal of Clinical Microbiology, 2005. v.43, n.9, p.4737-4743. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/16145135/ >. Accessed: Jan. 12, 2021. doi: 10.1128/JCM.43.9.4737-4743.2005.
https://pubmed.ncbi.nlm.nih.gov/16145135...
) and ST746 in Argentina (SREDNIK et al., 2018SREDNIK, M. E. et al. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. Journal of Dairy Research, 2018. v.85, n.1, p.57-63. Available from: <Available from: https://www.cambridge.org/core/journals/journal-of-dairy-research/article/abs/characterisation-of-staphylococcus-aureus-strains-isolated-from-mastitis-bovine-milk-in-argentina/4DE612151D432BD88F35458B03237533 >. Accessed: Jan. 23, 2021. doi: 10.1017/S0022029917000851.
https://www.cambridge.org/core/journals/...
). All ST746 S. aureus and most of the ST126 isolates deposited in PubMLST were isolated from bovine mastitis in Brazil, suggesting that these isolates may be adapted to Brazilian dairy herds (milk production system) and thereby easily spread among the properties, as mentioned above.
ST126 is a triple locus variant (TLV) and ST746 is a single locus variant (SLV) of ST97, which is the central genotype of CC97, considered a bovine-specific linage (BOSS et al., 2016BOSS, R. et al. Bovine Staphylococcus aureus: Subtyping, evolution, and zoonotic transfer. Journal of Dairy Science, 2016. v.99, n.1, p.515-528. Available from: <Available from: https://www.journalofdairyscience.org/article/S0022-0302(15)00834-6/fulltext >. Accessed: Jan. 23, 2021. doi: 0.3168/jds.2015-9589.
https://www.journalofdairyscience.org/ar...
; RABELLO et al., 2007RABELLO, R. F. et al. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. Journal of Medical Microbiology, 2007. v.56, n.11, p.1505-1511. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.47357-0#tab2 >. Accessed: Jan. 14, 2021. doi: 10.1099/jmm.0.47357-0.
https://www.microbiologyresearch.org/con...
; SAKWINSKA et al., 2011SAKWINSKA, O. et al. Staphylococcus aureus host range and human-bovine host shift. Applied and Environmental Microbiology, 2011. v.77, n.17, p.5908-5915. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3165375/ >. Accessed: Jan. 14, 2021. doi: 10.1128/AEM.00238-11.
https://www.ncbi.nlm.nih.gov/pmc/article...
). Although, transmission of CC97 isolates between cattle and humans is considered relatively rare, reports on human infections caused by isolates of this lineage are increasing (SPOOR et al., 2013SPOOR, L. E. et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio, 2013. v.4, n.4, p.1-6. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3747577/ >. Accessed: Jan. 22, 2021. doi: 10.1128/mBio.00356-13.
https://www.ncbi.nlm.nih.gov/pmc/article...
). Because of that, CC97 S. aureus has been considered an emerging cause of human infections, and the cows a potential reservoir for the emergence of new clones with the capacity for pandemic spread (SPOOR et al., 2013), although the epidemiological link is still unclear (MONECKE et al., 2011MONECKE, S. et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE, 2011. v.6, n.4. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3071808/ >. Accessed: Jan. 14, 2021. doi: 10.1371/journal.pone.0017936.
https://www.ncbi.nlm.nih.gov/pmc/article...
; SPOOR et al., 2013; SUNG et al., 2008SUNG, J. M. L.; LLOYD, D. H.; LINDSAY, J. A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology, 2008. v.154, n.7, p.1949-1959. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.2007/015289-0#tab2 >. Accessed: Jan. 22, 2021. doi: 10.1099/mic.0.2007/015289-0.
https://www.microbiologyresearch.org/con...
). A large number of isolates with MLST profiles genetically close to CC97 are of particular concern in Brazil since CC97 was already detected in isolates from samples of fresh Minas cheese (artisanal Brazilian cheese made using raw milk) (ALVES et al., 2018ALVES, V. F. et al. Molecular characterisation of Staphylococcus aureus from some artisanal Brazilian dairies. International Dairy Journal, 2018. v.85, p.247-253. Available from: <Available from: https://www.sciencedirect.com/science/article/abs/pii/S0958694618301547 >. Accessed: Jan. 16, 2021. doi: 10.1016/j.idairyj.2018.06.008.
https://www.sciencedirect.com/science/ar...
), a potential source for human disease caused by S. aureus. Furthermore, another transmission form that cannot be overlooked is from cows to farmworkers, who have constant and direct contact with potentially infected animals (SUNG et al., 2008).
Another relevant issue about CC97 is the emergence of MRSA among the isolates that belong to this clonal complex (BENTON et al., 2004BENTON, B. M. et al. Large-scale identification of genes required for full virulence of Staphylococcus aureus. Journal of Bacteriology, 2004. v.186, n.24, p.8478-8489. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC532413/ >. Accessed: Jan. 16, 2021. doi: 10.1128/JB.186.24.8478-8489.2004.
https://www.ncbi.nlm.nih.gov/pmc/article...
; FELTRIN et al., 2016FELTRIN, F. et al. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Applied and Environmental Microbiology, 2016. v.82, n.3, p.816-821. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4725266/ >. Accessed: Jan. 13, 2021. doi: 10.1128/AEM.02854-15.
https://www.ncbi.nlm.nih.gov/pmc/article...
; SPOOR et al., 2013SPOOR, L. E. et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio, 2013. v.4, n.4, p.1-6. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3747577/ >. Accessed: Jan. 22, 2021. doi: 10.1128/mBio.00356-13.
https://www.ncbi.nlm.nih.gov/pmc/article...
), which is a great public health problem due to zoonotic transmission. This fact draws attention to the two isolates oxacillin-resistant found in this study, both from Lima Duarte and belonging to the CC97. Although, disk-diffusion test using oxacillin is not the standard method to detect MRSA, it can be an indicator of resistance as it is used to detect methicillin resistance in other Staphylococcus species (CLSI, 2018cCLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Susceptibility Testing M100. 28th. ed. Wayne: Clinical and Laboratory Standards Institute , 2018c.). Also, beyond these two suggestive MRSA isolates, 26 isolates exhibited resistance to other penicillins [28/37 (75.68%)], most of the CC97 profile [24/29 (82.76%)], which highlighted the concern of zoonotic infections by isolates of this CC.
Despite the cow-to-cow transmission being the most common source of infections in bovine mastitis by S. aureus, the frequent occurrence of multiple isolates with low prevalence or incidence in infected herds suggested that this is not the only route of infection (RAINARD et al., 2017RAINARD, P. et al. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transboundary and Emerging Diseases, 2017. v.65, n.1, p.149-165. Available from: <Available from: https://onlinelibrary.wiley.com/doi/10.1111/tbed.12698 >. Accessed: Jan. 17, 2021. doi: 10.1111/tbed.12698.
https://onlinelibrary.wiley.com/doi/10.1...
). In this study, in addition to CC97, two other clonal complexes were observed, CC1 [3/37 (8.11%] and CC5 [2/37 (5.40%)]. These two clonal complexes are common and widespread, usually detected in human infections caused by S. aureus, but also described in mastitis cases worldwide (BUDD et al., 2015BUDD, K. E. et al. Extensive genomic diversity among bovine-adapted staphylococcus aureus: Evidence for a genomic rearrangement within CC97. PLoS ONE, 2015. v.10, n.8, p.1-17. Available from: <Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0134592 >. Accessed: Jan. 28, 2021. doi: 10.1371/journal.pone.0134592.
https://journals.plos.org/plosone/articl...
; CORTIMIGLIA et al., 2015CORTIMIGLIA, C. et al. Short communication: Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in Northern Italy. Journal of Dairy Science, 2015. v.98, n.4, p.2307-2311. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ >. Accessed: Jan. 17, 2021. doi: 10.3168/jds.2014-8923.
https://www.ncbi.nlm.nih.gov/pmc/article...
; HARAN et al., 2012HARAN, K. P. et al. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. Journal of Clinical Microbiology, 2012. v.50, n.3, p.688-695. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3295154/ >. Accessed: Jan. 21, 2021. doi: 10.1128/JCM.05214-11.
https://www.ncbi.nlm.nih.gov/pmc/article...
; MCMILLAN et al., 2016MCMILLAN, K. et al. Characterization of Staphylococcus aureus isolates from raw milk sources in Victoria, Australia. BMC Microbiology, 2016. v.16, n.1. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4966798/ >. Accessed: Jan. 17, 2021. doi: 10.1186/s12866-016-0789-1.
https://www.ncbi.nlm.nih.gov/pmc/article...
; SILVA et al., 2013SILVA, N. C. C. et al. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. Journal of Dairy Science, 2013. v.96, n.11, p.6856-6862. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/24054305/ >. Accessed: Jan. 17, 2021. doi: 10.3168/jds.2013-6719.
https://pubmed.ncbi.nlm.nih.gov/24054305...
; YANG et al., 2018YANG, X. et al. Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Frontiers in Microbiology, 2018. v.9, n.MAR, p.1-8. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5890145/ >. Accessed: Jan. 11, 2021. doi: 10.3389/fmicb.2018.00197.
https://www.ncbi.nlm.nih.gov/pmc/article...
; ZHANG et al., 2016ZHANG, L. et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microbial Pathogenesis, 2016. v.97, p.103-109. Available from: <Available from: http://dx.doi.org/10.1016/j.micpath.2016.06.005 >. Accessed: Jan. 14, 2021. doi: 10.1016/j.micpath.2016.06.005.
http://dx.doi.org/10.1016/j.micpath.2016...
). The evidence of cattle infected with S. aureus lineages commonly associated with human diseases draws attention to the role of human-to-bovine transmission in bovine mastitis. In fact, according to BOSS et al. (2016BOSS, R. et al. Bovine Staphylococcus aureus: Subtyping, evolution, and zoonotic transfer. Journal of Dairy Science, 2016. v.99, n.1, p.515-528. Available from: <Available from: https://www.journalofdairyscience.org/article/S0022-0302(15)00834-6/fulltext >. Accessed: Jan. 23, 2021. doi: 0.3168/jds.2015-9589.
https://www.journalofdairyscience.org/ar...
), S. aureus isolates from animal origin evolved from human-adapted isolates. In the MST analysis, comparing all Brazilian S. aureus isolates deposited in the PubMLST database and those from the present study (Figure 3b), it is observed great similarity among some isolates from milk and dairy products and isolates from human infections and environment, most belonging to CC1 and CC5, which also reinforce this epidemiological link. These findings underline the difficulty to control and eradicate S. aureus from the dairy production system, since the farmworkers may constitute a stable source of this pathogen. In this sense, specific-human and specific-bovine linages were observed at the same herd, suggesting that different reservoirs can be found in these farms, including humans (Table 3).
Furthermore, the isolates showed high resistance to penicillins and tetracycline (Table 1), which are commonly antimicrobials used for the treatment of mastitis in Brazilian dairy cattle, although there are no records of commercialization of veterinary drugs available. Regarding other antimicrobial groups tested, most of the isolates were susceptible, suggesting that these groups are not used as much as penicillins and tetracyclines to treat mastitis in this region. However, as 18.92% of the isolates were MDR, it is possible to suppose that these groups have been used as alternatives to penicillins and tetracyclines, which increases selective pressure and favors the emergence of MDR isolates and hinders mastitis control. In this context, the One Health strategy, which is based on the indivisible interrelation among human, animal, and environmental, should be considered in the surveillance of zoonotic pathogens, as S. aureus¸ mainly concerning antimicrobial resistance and foodborne diseases.
The isolates analyzed in this present study were isolated for over ten years. However, given the importance of S. aureus as a human and animal pathogen even today, information on epidemiological distribution, genetic evolution and adaptation to the host - as reported in this research - is of great importance to support and direct newer studies.
In conclusion, high genetic diversity and a great number of new STs and alleles were observed among S. aureus isolated from dairy cows in Minas Gerais, Brazil, besides genetic proximity between S. aureus isolates from humans and animal origin. Moreover, our results also showed high resistance to penicillin and tetracyclines, as well as suggestive of MRSA isolates, a potential threat to both animal and human health.
ACKNOWLEDGEMENTS
This study was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (Capes) (Finace Code 001), Fundação de Amparo à Pesquisa de Minas Gerais (Fapemig), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Empresa Brasileira de Pesquisa Agropecuária (Embrapa) - Gado de Leite. MS thanks Capes for her fellowship. MBH thanks CNPq for his fellowship.
REFERENCES
- AIRES-DE-SOUSA, M. et al. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Applied and Environmental Microbiology, 2007. v.73, n.12, p.3845-3849. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1932710/ >. Accessed: Jan. 15, 2021. doi: 10.1128/AEM.00019-07.
» https://doi.org/10.1128/AEM.00019-07.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1932710/ - ALVES, V. F. et al. Molecular characterisation of Staphylococcus aureus from some artisanal Brazilian dairies. International Dairy Journal, 2018. v.85, p.247-253. Available from: <Available from: https://www.sciencedirect.com/science/article/abs/pii/S0958694618301547 >. Accessed: Jan. 16, 2021. doi: 10.1016/j.idairyj.2018.06.008.
» https://doi.org/10.1016/j.idairyj.2018.06.008.» https://www.sciencedirect.com/science/article/abs/pii/S0958694618301547 - BENTON, B. M. et al. Large-scale identification of genes required for full virulence of Staphylococcus aureus. Journal of Bacteriology, 2004. v.186, n.24, p.8478-8489. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC532413/ >. Accessed: Jan. 16, 2021. doi: 10.1128/JB.186.24.8478-8489.2004.
» https://doi.org/10.1128/JB.186.24.8478-8489.2004.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC532413/ - BOSS, R. et al. Bovine Staphylococcus aureus: Subtyping, evolution, and zoonotic transfer. Journal of Dairy Science, 2016. v.99, n.1, p.515-528. Available from: <Available from: https://www.journalofdairyscience.org/article/S0022-0302(15)00834-6/fulltext >. Accessed: Jan. 23, 2021. doi: 0.3168/jds.2015-9589.
» https://doi.org/0.3168/jds.2015-9589.» https://www.journalofdairyscience.org/article/S0022-0302(15)00834-6/fulltext - BRITO, M. A. V. P. E; BRITO, J. R. F. Diagnóstico Microbiológico da Mastite. Juiz de Fora, MG: Embrapa Gado de Leite, 1999.
- BUDD, K. E. et al. Extensive genomic diversity among bovine-adapted staphylococcus aureus: Evidence for a genomic rearrangement within CC97. PLoS ONE, 2015. v.10, n.8, p.1-17. Available from: <Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0134592 >. Accessed: Jan. 28, 2021. doi: 10.1371/journal.pone.0134592.
» https://doi.org/10.1371/journal.pone.0134592.» https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0134592 - CLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 5th ed. CLSI standard VET01. 5th. ed. Wayne: Clinical and Laboratory Standards Institute, 2018.
- CLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals VET08. 4th. ed. Wayne: Clinical and Laboratory Standards Institute , 2018b.
- CLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). Performance Standards for Antimicrobial Susceptibility Testing M100. 28th. ed. Wayne: Clinical and Laboratory Standards Institute , 2018c.
- CORTIMIGLIA, C. et al. Short communication: Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in Northern Italy. Journal of Dairy Science, 2015. v.98, n.4, p.2307-2311. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ >. Accessed: Jan. 17, 2021. doi: 10.3168/jds.2014-8923.
» https://doi.org/10.3168/jds.2014-8923.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ - CUNHA, A. F. DA et al. Prevalence, etiology and risk factors of clinical mastitis in dairy cattle of Viçosa-MG. Acta Veterinaria Brasilica, 2016. v.10, n.1, p.48-54.
- ENRIGHT, M. C. et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology, 2000. v.38, n.3, p.1008-1015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ >. Accessed: Jan. 12, 2021. doi: 10.1128/JCM.38.3.1008-1015.2000.
» https://doi.org/10.1128/JCM.38.3.1008-1015.2000.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC86325/ - EWING, B. et al. Base-calling of automated sequencer races using phred. I. Accuracy assessment. Genome Research, 1998. v.8, n.3, p.175-85. Available from: <Available from: https://genome.cshlp.org/content/8/3/175.long >. Accessed: Jan. 16, 2021. doi: 10.1101/gr.8.3.175.
» https://doi.org/10.1101/gr.8.3.175.» https://genome.cshlp.org/content/8/3/175.long - FEIL, E. J. et al. How clonaliIs Staphylococcus aureus? Journal of Bacteriology, 2003. v.185, n.11, p.3307-3316. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC155367/ >. Accessed: Jan. 17, 2021. doi: 10.1128/JB.185.11.3307-3316.2003.
» https://doi.org/10.1128/JB.185.11.3307-3316.2003.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC155367/ - FELTRIN, F. et al. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Applied and Environmental Microbiology, 2016. v.82, n.3, p.816-821. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4725266/ >. Accessed: Jan. 13, 2021. doi: 10.1128/AEM.02854-15.
» https://doi.org/10.1128/AEM.02854-15.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4725266/ - FRANCISCO, A. P. et al. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics, 2009. v.10, p.1-15. Available from: <Available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-10-152 >. Accessed: Jan. 20, 2021. doi: 10.1186/1471-2105-10-152.
» https://doi.org/10.1186/1471-2105-10-152.» https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-10-152 - HARAN, K. P. et al. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. Journal of Clinical Microbiology, 2012. v.50, n.3, p.688-695. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3295154/ >. Accessed: Jan. 21, 2021. doi: 10.1128/JCM.05214-11.
» https://doi.org/10.1128/JCM.05214-11.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3295154/ - HEIKKILÄ, A. M. et al. Pathogen-specific production losses in bovine mastitis. Journal of Dairy Science, 2018. v.101, n.10, p.9493-9504. Available from: <Available from: http://dx.doi.org/10.3168/jds.2018-14824 >. Accessed: Jan. 14, 2021. doi: 10.3168/jds.2018-14824.
» https://doi.org/10.3168/jds.2018-14824.» http://dx.doi.org/10.3168/jds.2018-14824 - HUANG, X.; MADAN, A. CAP3: A DNA sequence assembly program. Genome Research, 1999. v.9, n.9, p.868-877. Available from: < Available from: https://www.genome.orgwww.genome.org >. Accessed: Jan. 18, 2021. doi: 10.1101/gr.9.9.868.
» https://doi.org/10.1101/gr.9.9.868.» https://www.genome.orgwww.genome.org - HUNTER, P. R.; GASTON, M. A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. Journal of Clinical Microbiology, 1988. v.26, n.11, p.2465-2466. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC266921/ >. Accessed: Jan. 11, 2021. doi: 10.1128/jcm.26.11.2465-2466.1988.
» https://doi.org/10.1128/jcm.26.11.2465-2466.1988.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC266921/ - KINROSS, P. et al. Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Surveillance and Outbreak Report, 2017. v.22, n.44, p.1-13. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5710135/ >. Accessed: Jan. 17, 2021. doi: 10.2807/1560-7917.ES.2017.22.44.16-00696.
» https://doi.org/10.2807/1560-7917.ES.2017.22.44.16-00696.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5710135/ - KULKARNI, A. G.; KALIWAL, B. B. Bovine mastitis: A review. International Journal of Recent Scientific Research, 2013. v.4, n.5, p.543-548. Available from: <Available from: http://www.recentscientific.com/bovine-mastitis-review >. Accessed: Jan. 11, 2021.
» http://www.recentscientific.com/bovine-mastitis-review - MAGIORAKOS, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 2011. v.18, n.3, p.268-281. Available from: <Available from: http://dx.doi.org/10.1111/j.1469-0691.2011.03570.x >. Accessed: Jan. 15, 2021. doi: 10.1111/j.1469-0691.2011.03570.x.
» https://doi.org/10.1111/j.1469-0691.2011.03570.x.» http://dx.doi.org/10.1111/j.1469-0691.2011.03570.x - MCMILLAN, K. et al. Characterization of Staphylococcus aureus isolates from raw milk sources in Victoria, Australia. BMC Microbiology, 2016. v.16, n.1. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4966798/ >. Accessed: Jan. 17, 2021. doi: 10.1186/s12866-016-0789-1.
» https://doi.org/10.1186/s12866-016-0789-1.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4966798/ - MESQUITA, A. A. et al. Staphylococcus aureus and streptococcus agalactiae: Prevalence, resistance to antimicrobials, and their relationship with the milk quality of dairy cattle herds in Minas Gerais state, Brazil. Pesquisa Veterinaria Brasileira, 2019. v.39, n.5, p.308-316. Available from: <Available from: https://www.scielo.br/j/pvb/a/4gysmHDfd3wqNh3YNtJnYBx/?lang=en >. Accessed: Jan. 16, 2021. doi: 10.1590/1678-5150-PVB-5821.
» https://doi.org/10.1590/1678-5150-PVB-5821.» https://www.scielo.br/j/pvb/a/4gysmHDfd3wqNh3YNtJnYBx/?lang=en - MONECKE, S. et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE, 2011. v.6, n.4. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3071808/ >. Accessed: Jan. 14, 2021. doi: 10.1371/journal.pone.0017936.
» https://doi.org/10.1371/journal.pone.0017936.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3071808/ - PÉREZ, V. K. C. et al. Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. Journal of Global Antimicrobial Resistance, 2020. v.22, p.792-802. Available from: <Available from: https://doi.org/10.1016/j.jgar.2020.06.010 >. Accessed: Jan. 17, 2021. doi: 10.1016/j.jgar.2020.06.010.
» https://doi.org/10.1016/j.jgar.2020.06.010.» https://doi.org/10.1016/j.jgar.2020.06.010 - RABELLO, R. F. et al. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. Journal of Medical Microbiology, 2007. v.56, n.11, p.1505-1511. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.47357-0#tab2 >. Accessed: Jan. 14, 2021. doi: 10.1099/jmm.0.47357-0.
» https://doi.org/10.1099/jmm.0.47357-0.» https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.47357-0#tab2 - RAINARD, P. et al. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transboundary and Emerging Diseases, 2017. v.65, n.1, p.149-165. Available from: <Available from: https://onlinelibrary.wiley.com/doi/10.1111/tbed.12698 >. Accessed: Jan. 17, 2021. doi: 10.1111/tbed.12698.
» https://doi.org/10.1111/tbed.12698.» https://onlinelibrary.wiley.com/doi/10.1111/tbed.12698 - SAKWINSKA, O. et al. Staphylococcus aureus host range and human-bovine host shift. Applied and Environmental Microbiology, 2011. v.77, n.17, p.5908-5915. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3165375/ >. Accessed: Jan. 14, 2021. doi: 10.1128/AEM.00238-11.
» https://doi.org/10.1128/AEM.00238-11.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3165375/ - SAUNDERS, N. A.; HOLMES, A. Multilocus sequence typing (MLST) of Staphylococcus aureus. Methods in Molecular Biology, 2007. v.391, p.71-85. Available from: <Available from: https://link.springer.com/protocol/10.1007%2F978-1-59745-468-1_6 >. Accessed: Jan. 18, 2021. doi: 10.1007/978-1-59745-468-1_6.
» https://doi.org/10.1007/978-1-59745-468-1_6.» https://link.springer.com/protocol/10.1007%2F978-1-59745-468-1_6 - SILVA, N. C. C. et al. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. Journal of Dairy Science, 2013. v.96, n.11, p.6856-6862. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/24054305/ >. Accessed: Jan. 17, 2021. doi: 10.3168/jds.2013-6719.
» https://doi.org/10.3168/jds.2013-6719.» https://pubmed.ncbi.nlm.nih.gov/24054305/ - SMITH, E. M. et al. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. Journal of Clinical Microbiology, 2005. v.43, n.9, p.4737-4743. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/16145135/ >. Accessed: Jan. 12, 2021. doi: 10.1128/JCM.43.9.4737-4743.2005.
» https://doi.org/10.1128/JCM.43.9.4737-4743.2005.» https://pubmed.ncbi.nlm.nih.gov/16145135/ - SOBRAL, D. et al. High throughput multiple locus variable number of tandem repeat analysis (MLVA) of Staphylococcus aureus from human, animal and food sources. PLoS ONE, 2012. v.7, n.5, p.e33967. Available from: <Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033967 >. Accessed: Jan. 17, 2021. doi: 10.1371/journal.pone.0033967.
» https://doi.org/10.1371/journal.pone.0033967.» https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033967 - SPOOR, L. E. et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio, 2013. v.4, n.4, p.1-6. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3747577/ >. Accessed: Jan. 22, 2021. doi: 10.1128/mBio.00356-13.
» https://doi.org/10.1128/mBio.00356-13.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3747577/ - SREDNIK, M. E. et al. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. Journal of Dairy Research, 2018. v.85, n.1, p.57-63. Available from: <Available from: https://www.cambridge.org/core/journals/journal-of-dairy-research/article/abs/characterisation-of-staphylococcus-aureus-strains-isolated-from-mastitis-bovine-milk-in-argentina/4DE612151D432BD88F35458B03237533 >. Accessed: Jan. 23, 2021. doi: 10.1017/S0022029917000851.
» https://doi.org/10.1017/S0022029917000851.» https://www.cambridge.org/core/journals/journal-of-dairy-research/article/abs/characterisation-of-staphylococcus-aureus-strains-isolated-from-mastitis-bovine-milk-in-argentina/4DE612151D432BD88F35458B03237533 - SUNG, J. M. L.; LLOYD, D. H.; LINDSAY, J. A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology, 2008. v.154, n.7, p.1949-1959. Available from: <Available from: https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.2007/015289-0#tab2 >. Accessed: Jan. 22, 2021. doi: 10.1099/mic.0.2007/015289-0.
» https://doi.org/10.1099/mic.0.2007/015289-0.» https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.2007/015289-0#tab2 - TIBAYRENC, M. Microbial molecular epidemiology : An overview. In: CAUGANT, D. A. (Org.). Molecular Epidemiology of Microorganisms. 1. ed. Totowa, New Jersey: Humana Press, 2009, p. 1-12.
- VIGUIER, C. et al. Mastitis detection: current trends and future perspectives. Trends in Biotechnology, 2009. v.27, n.8, p.486-493. Available from: <Available from: https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(09)00112-7 >. Accessed: Jan. 14, 2021. doi: 10.1016/j.tibtech.2009.05.004.
» https://doi.org/10.1016/j.tibtech.2009.05.004» https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(09)00112-7 - VLIEGHER, S. DE et al. Invited review: Mastitis in dairy heifers: Nature of the disease, potential impact, prevention, and control. Journal of Dairy Science, 2012. v.95, n.3, p.1025-1040. Available from: <Available from: http://dx.doi.org/10.3168/jds.2010-4074 >. Accessed: Jan. 11, 2021. doi: 10.3168/jds.2010-4074.
» https://doi.org/10.3168/jds.2010-4074.» http://dx.doi.org/10.3168/jds.2010-4074 - YANG, X. et al. Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Frontiers in Microbiology, 2018. v.9, n.MAR, p.1-8. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5890145/ >. Accessed: Jan. 11, 2021. doi: 10.3389/fmicb.2018.00197.
» https://doi.org/10.3389/fmicb.2018.00197.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5890145/ - ZHANG, L. et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microbial Pathogenesis, 2016. v.97, p.103-109. Available from: <Available from: http://dx.doi.org/10.1016/j.micpath.2016.06.005 >. Accessed: Jan. 14, 2021. doi: 10.1016/j.micpath.2016.06.005.
» https://doi.org/10.1016/j.micpath.2016.06.005.» http://dx.doi.org/10.1016/j.micpath.2016.06.005
-
CR-2021-0643.R2
Edited by
Publication Dates
-
Publication in this collection
24 June 2022 -
Date of issue
2023
History
-
Received
01 Sept 2021 -
Accepted
15 Mar 2022 -
Reviewed
18 May 2022