Abstract
The second-generation bioethanol employs lignocellulosic materials degraded by microbial cellulases in their production. The fungus Trichoderma reesei is one of the main microorganisms producing cellulases, and its genetic modification can lead to the optimization in obtaining hydrolytic enzymes. This work carried out the deletion of the sequence that encodes the zinc finger motif of the transcription factor ACE1 (cellulase expression repressor I) of the fungus T. reesei RUT-C30. The transformation of the RUT-C30 lineage was confirmed by amplification of the 989 bp fragment relative to the selection marker, and by the absence of the zinc finger region amplification in mutants, named T. reesei RUT-C30Δzface1. The production of cellulases by mutants was compared to RUT-C30 and measured with substrates carboxymethylcellulose (CMC), microcrystalline cellulose (Avicel®) and Whatman filter paper (PF). The results demonstrated that RUT-C30Δzface1 has cellulolytic activity increased 3.2-fold in Avicel and 2.1-fold in CMC and PF. The mutants presented 1.4-fold higher sugar released in the hydrolysis of the biomass assays. These results suggest that the partial deletion of ace1 gene is an important strategy in achieving bioethanol production on an industrial scale at a competitive price in the fuel market.
Keywords:
Trichoderma reesei; zinc finger motif deletion; ACE1; cellulase; bioethanol
INTRODUCTION
On average, 80% of the energy generated in the world comes from fossil fuels; however, these sources of energy are not renewable, and their reserves are rapidly depleting. Predictions estimate that in some decades fossil fuel reserves will suffer total depletion [11 Gupta A, Verma JP. Sustainable bio-ethanol production from agro-residues: a review. Renew Sust Energ Rev. 2015 Jan; 41:550-67.
2 Popp J, Lakner Z, Harangi-Rákos M, Fári M. The effect of bioenergy expansion: food, energy, and environment. Renew Sust Energ Rev. 2014 Apr; 32:559-78.-33 Raghuwanshi S, Deswal D, Karp M, Kuhad RC. Bioprocessing of enhanced cellulase production from a mutant of Trichoderma asperellum RCK2011 and its application in hydrolysis of cellulose. Fuel (Lond). 2014 May; 124:183-9.]. Biomass is the most abundant carbon source in the world and, therefore, has become one of the main sources of energy produced and consumed in the world [4Santos FA, Queiróz JD, Colodette JL, Fernandes SA, Guimarães VM, Rezende ST. [Potential of sugarcane straw for ethanol production]. Quim Nova. 2012 Jan; 35(5):1004-10. Portuguese.4].
The term “second generation fuels” refers to the production of ethanol from lignocellulosic biomass [55 Chiaramonti D, Prussi M, Ferrero S, Oriani L, Ottonello P, Torre P, et al. Review of pretreatment processes for lignocellulosic ethanol production, and development of an innovative method. Biomass Bioenerg. 2012 Nov; 46:25-35.], which derived from plant residues and the production of first generation ethanol, crops intended for the production of bioethanol, forest residues, industrial waste, or solid waste [66 Gomes D, Rodrigues AC, Domingues L, Gama M. Cellulase recycling in biorefineries-is it possible? Appl Microb Biotechnol. 2015 May; 99(10):4131-43.,7Imamoglu E, Sukan FV. Scale-up and kinetic modeling for bioethanol production. Bioresour Technol. 2013 Sept; 144:311-20.7]. However, there are relevant obstacles that need to be overcome, such as the cost of production, technological problems, and environmental problems [88 Ruiz HA, Silva DP, Ruzene DS, Lima LF, Vicente AA, Teixeira JA. Bioethanol production from hydrothermal pretreated wheat straw by a flocculating Saccharomyces cerevisiae strain-Effect of process conditions. Fuel. 2012 May; 95:528-36.].
In order to avoid uncontrolled expansion of cultivated areas, biotechnological techniques have been employed to allow the use of residual lignocellulosic biomass from sugarcane to produce bioethanol [88 Ruiz HA, Silva DP, Ruzene DS, Lima LF, Vicente AA, Teixeira JA. Bioethanol production from hydrothermal pretreated wheat straw by a flocculating Saccharomyces cerevisiae strain-Effect of process conditions. Fuel. 2012 May; 95:528-36.].
Second generation ethanol employs a more complex substrate, which is not easily accessible for microbial fermentation. The lignocellulosic materials consist mainly of cellulose (40-60% of the total dry weight), hemicellulose (20-40%), and lignin (10-25%) [66 Gomes D, Rodrigues AC, Domingues L, Gama M. Cellulase recycling in biorefineries-is it possible? Appl Microb Biotechnol. 2015 May; 99(10):4131-43.,99 Baeyens J, Kang Q, Appels L, Dewil R, Lv Y, Tan T. Challenges and opportunities in improving the production of bio-ethanol. Prog Energy Combust Sci. 2015 Apr; 47:60-88.].
The lignocellulose is processed for bioethanol production through three major operations: (1) pretreatment (i.e., the delignification necessary to release cellulose and hemicellulose before hydrolysis), (2) cellulose and hemicellulose hydrolysis (i.e., the production of fermentable sugars, such as glucose, xylose, arabinose, galactose, and mannose), and (3) reducing sugars fermentation [88 Ruiz HA, Silva DP, Ruzene DS, Lima LF, Vicente AA, Teixeira JA. Bioethanol production from hydrothermal pretreated wheat straw by a flocculating Saccharomyces cerevisiae strain-Effect of process conditions. Fuel. 2012 May; 95:528-36.,1010 Sarkar N, Ghosh SK, Bannerjee S, Aikat K. Bioethanol production from agricultural wastes: An overview. Renew Energy. 2012 Jan; 37(1):19-27.]. The enzymatic hydrolysis process has been performed by microorganisms that have cellulolytic enzymes secretion ability [11 Gupta A, Verma JP. Sustainable bio-ethanol production from agro-residues: a review. Renew Sust Energ Rev. 2015 Jan; 41:550-67.]. Cellulolytic microorganisms, like bacteria and filamentous fungi, produce a set of enzymes that synergically hydrolyze crystalline cellulose to small oligosaccharides and to glucose [1111 Sukumaran RK, Singhania RR, Pandey A. Microbial cellulases-production, applications and challenges.J Sci Ind Res (India). 2005 Nov; 64(11):832-44.]. Biological hydrolysis can reduce approximately 40% of the production cost of ethanol from biomass [1212 Bendig C, Weuster-Botz D. Reaction engineering analysis of cellulase production with Trichoderma reesei RUT-C30 with intermittent substrate supply. Bioprocess Biosyst Eng. 2013 Jul; 36(7):893-900.].
T. reesei is a mesophilic filamentous fungus that quickly became one of the most important fungi for biotechnology, due to its great capacity to produce a large number of cellulolytic enzymes [1313 Peterson R, Nevalainen H. Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology. 2012 Jan; 158(1):58-68.]. T. reesei genome comprises 9,143 genes [1414 Kubicek CP. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J Biotechnol. 2013 Jan; 163(2):133-42.]. Five important transcription factors that regulates cellulases and hemicellulases synthesis have been described as XYR1, ACE2, and the 2/3/5 HAP complex-like positive regulators and negative regulators, such as ACE1 and CRE1 (catabolic carbon repressor) [1515 Castro LS, Antoniêto AC, Pedersoli WR, Silva-Rocha R, Persinoti GF, Silva RN. Expression pattern of cellulolytic and xylanolytic genes regulated by transcriptional factors XYR1 and CRE1 are affected by carbon source in Trichoderma reesei. Gene Expr Patterns. 2014 Mar; 14(2):88-95.]. ACE1 contains three Cys2His2-type zinc finger sequences and is capable to binding in vitro to 8 sites containing 5 'AGGCA sequence over a 1.15 kb cellobiohydrolase 1 (cbh1) promoter [1616 Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003 Jan; 69(1):56-65.].
Natick laboratories were the pioneers in the generation of mutants from T. reesei. QM6a, the mutant QM9414, was the most successful and presented cellulase production up to four times greater than the wild-type lineage. From the line, QM9414 was created by the hypercellulolytic mutant RUT-C30, which is the most reported and used mutant strain of all in industrial processes involving the hydrolysis of biomass [1313 Peterson R, Nevalainen H. Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology. 2012 Jan; 158(1):58-68.].
The objective of this work was to transform the RUT-C30 line by deleting the sequence that encodes the zinc finger of the repressor transcription factor cellulase ACE1 to obtain an optimized cell line for the production of cellulase.
MATERIAL AND METHODS
Strains and Plasmids
The target of experiments was the T. reesei RUT-C30 strain, which was stored in a malt extract agar medium (MEX - 0.3% malt extract, 0.5% mycological peptone, 1.5% agar and 1% glucose with pH 5.4) until 15 days for the strain maintenance. For deletion cassette construction were used the pRS426 vector [1717 Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992 Jan; 110(1):119-22.], the hph selection marker (which confers resistance to hygromycin B), the 5' and 3' flanking regions of the encoding sequence of the zinc finger motif of the cellulase repressor gene ace1 and S. cerevisiae SC9721. The yeast strain was stored in YPD medium (1% yeast extract, 2% peptone, and 2% glucose).
Primers Construction and Obtaining of ace1 5’ and 3' Regions and hph Selection Marker
In order to promote only the deletion of zinc finger motifs region in ACE1 transcription factor, specific primers were generated for flanking sequences of this gene region. First, the T. reesei ace1 gene sequence (accession number 75418 deposited at the Joint Genome Institute (JGI) website: http://genome.jgi-psf.org/) was translated using the BioEdit® program (version 7.1.3.0). Subsequently the zinc finger motifs sequence (Tyr, Phe)-X-Cys-X2-5-Cys-X3-(Tyr, Phe)-X5-Leu-X2-His-X3-5-His was searched for primers construction. Specific complementary sequences were added to the primers for 5' and 3' regions flanking the target sequence and the hph selection marker, according to the Schuster-generated database [1818 Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, et al. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels 2012 Jan; 5(1):1-10.] (Table I). The 5' and 3' regions were amplified from the T. reesei QM6a fungus, and the hph was amplified from pΔku70 (provided by Dr. Roberto Nascimento Silva). The mixture PCR contained 2.5 μL of Taq DNA polymerase buffer (Invitrogen), 0.5 μL of phosphate deoxyribonucleotides 10 mM, 1.0 μL of MgCl2 25 mM, 0.5 μL of Primer Forward and Reverse 100 pmol/μL, 1 μL of DNA sample 0.2 μg/μL, 1.0 μL of High Fidelity® Taq DNA polymerase (Invitrogen), and 18.5 μL of Milli-Q nuclease free water. The protocol was: 1 cycle of 95°C for 3 min; 35 cycles of 94 °C for 30 s; 60 °C for 30 s; 72 °C for 2 min; and a final extension at 72 °C for 10 min. The amplified sequences were purified using a PureLink® kit (Invitrogen), following the manufacturer's guidelines.
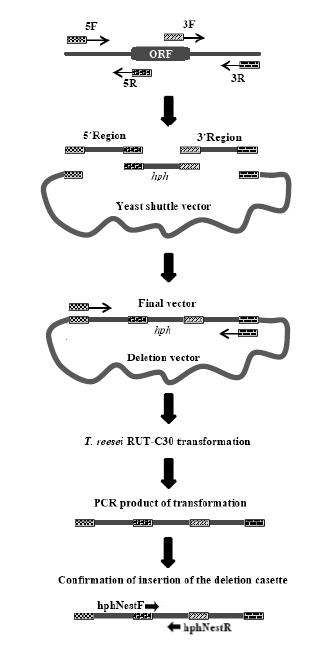
Deletion Cassette Construction and Yeast Mediated Recombination
The methodology used for the deletion cassette construction was adapted from Mota Júnior and Colot and coauthors [1919 Mota Júnior AO, Malavazi I, Soriani FM, Heinekamp T, Jacobsen I, Brakhage AA, et al. Molecular characterization of the Aspergillus fumigatus NCS-1 homologue, NcsA. Mol Genet Genomics 2008 Dec; 280(6):483-95.,2020 Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006 Jul; 103(27):10352-7.] (Figure 1). Yeast pre-inoculum was prepared in 10 mL of YPD medium, which was incubated for 16 h at 30°C under 200 rpm orbital shaking. The following day, 2.5 mL of this pre-inoculum was inoculated into 100 mL of YPD medium and incubated for 4 h under the same conditions described before. Subsequently, the inoculum was centrifuged at 5000 g for 5 min, the supernatant was rejected, the pellet was resuspended in 20 mL of sterile water, and the centrifugation process was repeated. The precipitate was resuspended with 1 mL of lithium acetate, 1 M, and TE buffer (100 mM Tris-HCl, 100 μM EDTA) solution.
After, 100 μL of the competent cells were added to Mix 1, which contained 200 ng of both 5' and 3' fragments flanking the target sequence, 100 ng of selection marker (hph), 100 μg salmon sperm, and 100 ng of linearized pRS426 vector through the digestion reaction with restriction enzymes EcoRI and XhoI. Also added in this mixture was 600 μL of Mix 2 (i.e., 800 μL of polyethylene glycol 3550 50%, 100 μL of lithium acetate 1 M, and 100 μL of Milli-Q water). Homogenization was performed by inversion, and the mixture was incubated at 30°C for 30 min under 200 rpm shaking. In addition, 70 μL of dimethyl sulfoxide (DMSO) was added, and the mixture was homogenized by inversion and incubated at 42°C for 15 min, followed by 2 minutes in an ice bath. Afterwards, 700 μL of Milli-Q water was added and mixed by inversion. Another centrifugation was performed for 30 s at 6080 g. It discarded 800 μL of the supernatant, and the remainder was plated in SC-URA medium (0.7% YNB [Yeast Nitrogen Base without amino acids], 2% glucose, 1.7% agar, and the following amino acids: 0.01% leucine, 0.01% lysine, 0.01% tryptophan 0.01%, and 0.005% histidine) with Drigalski loop aid. The incubation was performed at 30°C for three to four days until growth of visible yeast colonies. The experiment controls were as follows: negative control I (transformation only with water and yeast); negative control II (transformation with open vector and salmon sperm); and positive control (transformation with closed vector and salmon sperm).
Methodology for the yeast genomic DNA extraction after the deletion cassette construction was followed, as described by Mota Júnior [1919 Mota Júnior AO, Malavazi I, Soriani FM, Heinekamp T, Jacobsen I, Brakhage AA, et al. Molecular characterization of the Aspergillus fumigatus NCS-1 homologue, NcsA. Mol Genet Genomics 2008 Dec; 280(6):483-95.]. The final deletion cassette was obtained through a PCR, using High Fidelity® Taq DNA polymerase (Invitrogen), yeast genomic DNA, and ace1hph5F and ace1hph3R primers, according to the conditions described above with modifications in the 4 min extension time.
Schematic representation of yeast-mediated S. cerevisiae vector construction and confirmation of deletion cassette insertion, as modified from Schuster and coauthors [1818 Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, et al. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels 2012 Jan; 5(1):1-10.].
T. reesei RUT-C30 Transformation
After the deletion cassette assembly, the T. reesei RUT-C30 strain protoplasts were transformed according to Schuster and coauthors [1818 Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, et al. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels 2012 Jan; 5(1):1-10.]. After transformation, the visible growth transformants were transferred to several plates with MEX medium and selection reagent (100 μg/mL hygromycin) and incubated at 30°C until sporulation. From these plates, a spore solution was prepared and inoculated in MEX medium plus Triton X-100 0.1% to obtain isolated colonies. The cultured colonies were inoculated on plates with MEX medium with selection reagent and incubated at 30°C until sporulation.
T. reesei RUT- C30 ACE1 Deletion Confirmation
First, the genomic DNA of each strain was extracted according to the methodology adapted from Cassago and coauthors, Doyle, and Plaza and coauthors [21Cassago A, Panepucci R, Baião A, Henrique-Silva F. Cellophane based mini-prep method for DNA extraction from the filamentous fungus Trichoderma reesei. BMC Microbiol. 2002 Jun; 2,14-8.21
22 Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19(1):11-5.-2323 Plaza GA, Upchurch R, Brigmon RL, Whitman WB, Ulfig K. Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Pol J Environ Stud. 2004 Jan; 13(3):315-8.]. The fungus was grown in MEX medium, incubated at 30°C and 150 rpm for 24 h. Approximately 0.5 mL of the mycelium, 600 μL of the extraction solution (0.05 M EDTA; 1% SDS) and 1 g of glass beads 212-300 (m (Sigma() were transferred to a 1.5 mL microtube and mixed in the Vortex-Genie2® for 10 min, followed by incubation at 66°C for 20 min. The sample was centrifuged at 16,000 g for 10 min at room temperature, and 400 μL of the supernatant was transferred to a new microtube.
Then, 50 μL of RNase 20 mg/mL was added by inverting the mixture and followed by incubation at 37°C for 60 min at 500 rpm. It was added to 300 μL of phenol, mixed by inversion, and followed by centrifugation at 16,000 g for 10 min. Next, 400 μL of the supernatant was transferred to a new microtube, and 400 μL of chloroform was added: isoamyl alcohol (24:1), followed by centrifugation at 16,000 g for 5 min. This step was repeated twice and 400 μL of the supernatant was transferred to a microtube with 63 μL of 5 M potassium acetate (pH 4.8) and mixed by inversion. The samples were incubated at 2°C for 35 min and centrifugated at 17,320 g for 5 min at 4°C. Then, 400 μL of the supernatant was transferred to a new microtube, and 1 mL of 100% ice cold ethanol was mixed by inversion, followed by centrifugation at 17,320 g for 5 min at 4°C. The supernatant was discarded, and the sample was washed twice with 500 μL of 70% ice cold ethanol. After the sample dried, the precipitate was resuspended in 50 μL of TE/RNase buffer.
The T. reesei strain transformation was verified by PCR performed with oligonucleotides hphNestF and hphNestR for hph gene amplification (see Table 1). The PCR mixture contained 12.5 μL of GoTaq® Long PCR Master Mix (Promega), 0.5 μL of each primer (100 pmol/μL), 1.0 μL of genomic DNA (150 ng/μL), and 10.5 μL of Milli-Q water, at the following temperatures: 1 cycle of 95 °C for 3 min; 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min and 30 s; followed by a final extension of 10 min at 72 °C. A second reaction for confirmation was performed with the oligonucleotides ace1SC and ace1ZFSCR for amplification of the zinc finger coding region (see Table 1). The PCR reaction contained 1.0 μL of GoTaq® (Promega), 1.0 μL of each primer (100 pmol/μL), 1.0 μL of genomic DNA (100 ng/μL), 5.0 μL of 10X Taq Buffer, 1.0 μL dNTP (10 mM), and 40.0 μL Milli-Q Water, at the following temperatures: 1 cycle of 96°C for 3 min, 35 cycles of 95 °C for 30 s, 48°C for 40 s, and 72 °C for 1 min, followed by 1 cycle of 10 min at 72 °C.
Growth Rate of Strains
The growth characteristics was observed after inoculum of the strains on MEX agar medium, supplemented with 1% glucose and incubation at 28°C by 144 h. The colonies’ growth was measured every 24 h, and the results from RUT-C30Δzface1-1, RUT-C30Δzface1-2, and RUT-C30Δzface1-3 were compared with the RUT-C30 strain.
Enzymatic Production
A methodology adapted from Ahamed and Vermette [2424 Ahamed A, Vermette P. Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochem Eng J. 2008 Jul; 40(3):399-407.] was used for enzymatic production. The transformed RUT-C30 T. reesei cultures were inoculated in a 125 mL Erlenmeyer flask with 20 mL culture medium for cellulase production (1% cellulose, 1% yeast extract, 1% glucose, 0.03% MgSO4.7H2O, 0.2% KH2PO4, 0.14% (NH4)2SO4, 0.04% CaCl2, and 4 mL element traces solution 50X), following incubation at 30°C under orbital shaking at 150 rpm for 24 h. Afterwards, 5 mL of medium was added for cellulase induction with 25% lactose, without glucose and cellulose. It was incubated again under the same conditions for 24 h. The cultures obtained from the liquid media were vacuum filtered on through a Büchner funnel and Whatman Grade 1 filter paper, yielding a cell-free filtrate and used as enzymatic extract to perform the determination of activities of cellulase and other glycoside hydrolases (i.e., xylanase and pectinase).
Enzyme Assays
The cellulase activities of endoglucanase, exoglucanase, and total cellulase were performed using carboxymethylcellulose (CMC), microcrystalline cellulose (Avicel®) and Whatman (PF) filter paper as substrates, respectively. The activities of pectinase, invertase, and xylanase were performed using pectin, sucrose, and xylan, respectively, according to the methodology described by Miller [2525 Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959 Mar; 31(3):426-8.] - using a 3,5-dinitrosalicylic acid reagent (DNS). The system was standardized by a glucose, galacturonic acid, and xylose calibration curves of 0.1 to 1.0 mg/mL. One enzymatic activity unit was defined as the enzyme amount that release 1 μmol of reducing sugar per minute under the assay conditions.
Hydrolysis Tests
Enzymatic Index
The enzymatic index was measured by plate hydrolysis tests, according to the methodology adapted from Florencio and coauthors [2626 Florencio C, Couri S, Farinas CS. Correlation between Agar Plate Screening and Solid-State Fermentation for the Prediction of Cellulase Production by Trichoderma Strains. Enzyme Res. 2012 Nov; 2012:1-7.]. The strains were inoculated for 96 h at 28 °C in Petri dishes containing solid MEX medium and supplemented with 2% glucose and 1% CMC. After 96 h, 10 mL of Congo red solution (2.5 g/L) was added and incubated at room temperature for 15 min. After, the cultures were incubated with NaCl 1 mol/L for 15 min. After formation of the hydrolysis halo, the enzymatic index was calculated by measuring the diameter of the hydrolysis halo and the size of the colonies.
Biomass Hydrolysis
The biomass hydrolysis efficiency by enzymatic extract of each mutant was carried out according to the adapted methodology described by Ribeiro and coauthors [2727 Ribeiro LF, de Lucas RC, Vitcosque GL, Ribeiro LF, Ward RJ, Rubio MV, et al. A novel thermostable xylanase GH10 from Malbranchea pulchella expressed in Aspergillus nidulans with potential applications in biotechnology. Biotechnol Biofuels 2014 Jul; 7(1):115-126.] and compared to enzymatic extract from RUT-C30 strain. In addition, we used 20 mg ground sugarcane bagasse, 0.2 mg extract protein and sufficient amount of sodium acetate buffer 100 mM pH 5.0 for a 1.5 mL. The samples were incubated at 30 °C and aliquots were withdrawn at 24 h, followed by boiling for 10 min. Subsequently, the samples were centrifuged for 10 min at 10,000 g, and the supernatant was used for reducing sugar determination according to Miller [2525 Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959 Mar; 31(3):426-8.].
Results Reproducibility and Statistical Analysis
The data were submitted to analysis of variance (ANOVA) in the GraphPad Prisma® 6.0, and the means were compared by the Bonferroni's or Dunnett's tests when significant differences were observed.
RESULTS
Construction of T. reesei RUT-C30 Deletion Mutants
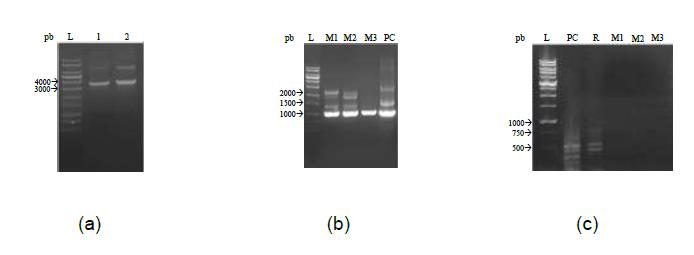
The deletion cassette was assembled by recombination and mediated by S. cerevisiae (SC9721) yeast using the purified 5' and 3' region fragments, purified hph selection marker, and previously digested and purified plasmid pRS426. After recombination, the yeast genomic DNA was used for the deletion cassette amplification. Figure 2 A shows the predicted PCR product with 3501 pb, which was purified and used in T. reesei RUT-C30 transformation assays.
After the T. reesei RUT-C30 strain transformation with the ace1 deletion cassette, the genomic DNA was extracted in order to confirm the deletion. First, the insertion of the deletion cassette was verified on three mutants by amplification of the hph selection marker (Figure 2 B), which were named RUT-C30Δzface1-1, RUT-C30Δzface1-2, and RUT-C30Δzface1-3. The observed fragment was 989 pb, which confirms the presence of the selection marker. After, the deletion of the sequence encoding the zinc finger of ACE1 was confirmed by the absence of amplification of the region containing the zinc finger sequence in the three mutants, whereas the RUT-C30 and QM9414 strains showed amplification of a fragment with 479 pb (see Figure 2C).
(A) Deletion cassette amplification. Horizontal electrophoresis by 0.7% agarose gel with amplification of the fragment with the hph deletion cassette with 3,500 base pairs. Symbols: (L) DNA Ladder 1Kb Ludwig® Biotec; (1) sample 1 and (2) sample 2. (B) Confirmation of deletion cassette insertion into the transformed T. reesei strains. Horizontal electrophoresis by 0.7% agarose gel, in which amplification of a fragment of the hph selection marker region is observed with 989 base pairs. Symbols: (L) DNA Ladder 1 Kb Ludwig® Biotec; (M1) RUT-C30Δzface1-1; (M2) RUT-C30Δzface1-2; (M3) RUT-C30Δzface1-3 and (PC) positive reaction control (hph). (C) Confirmation of the deletion of the zinc finger encoding region of the ACE1 transcription factor. Horizontal electrophoresis by 1.5% agarose gel, demonstrating the amplification of a fragment of the coding region of the zinc finger, with 479 base pairs for the positive control and parental lineage. Because there was a deletion on the mutant strains, there was no amplification. Symbols: (L) DNA Ladder 1kb RTU (Reading-to-use), (PC) Positive Control, (R) RUT-C30, (M1) RUT-C30∆zface1-1, (M2) RUT-C30∆zface1-2 e (M3) RUT-C30∆zface1-3.
Lineage Growth Rate
After 24 h of incubation, only the mutant strains showed growth. After 96 h of incubation, the mutants had a mycelial size of 2.54 times greater than the RUT-C30 strain. At 120 h, there was a significant difference between the mycelial sizes of the mutants and the parental strain. The RUT-C30 exceeded the size of the mutant strains only after 144 h of incubation (Figure 3).
The superior growth of the mutant strains on initial incubation time can leave to an increase of the enzymatic secretion in a short period of time. Thus, the next assay investigated the enzymatic production after 24 h of induction.
Mycelial growth of mutants, relative to RUT-C30. The colonies of the three mutant and parental lines were measured in millimeters every 24 hours for a total of 144 hours of incubation at 28°C. The statistical analysis was performed according to the Bonferroni's test using significance level of p < 0.05, in which (**) p < 0.01, (***) p < 0.001 and (****) p < 0.0001. Error bars represent the standard deviation of the results for each variable.
Enzymatic Production
The results after 24 h of enzymatic induction with lactose show an increase in the cellulases and pectinases production by the mutant strains, when compared to the RUT-C30. The production of exoglucanase was greater by mutants RUT-C30Δzface1-1 and RUT-C30Δzface1-3, which show 3.2 and 2.0-fold higher, respectively, than RUT-C30 (Figure 4). The production of total cellulase also showed an increase of 2.1-fold for RUT-C30Δzface1-1, 1.6-fold for RUT-C30Δzface1-2, and 1.8-fold for RUT-C30Δzface1-3. The RUT-C30Δzface1-1, RUT-C30Δzface1-2 and RUT-C30Δzface1-3 mutants presented endoglucanase production of 1.7, 2.0, and 2.1-fold higher than RUT-C30, respectively.
In addition to the optimization of the secretion of all cellulases, it was possible to observe an increase of pectinase production. The mutants RUT-C30Δzface1-1, RUT-C30Δzface1-2, and RUT-C30Δzface1-3 showed an increase of 1.9, 2.4, and 1.3-fold, in relation to RUT-C30.
Comparison of the enzymatic production in 24 hours of lactose induction. Statistical analysis performed according to Dunnett's test using significance level of p < 0.05, in which (**) p < 0.01 and (****) p < 0.0001. Error bars represent the standard deviation of the results for each variable.
Hydrolysis Tests
Enzymatic Index and Biomass Hydrolysis
All mutant strains showed a higher hydrolysis index than RUT-C30, after growth by 96 h in Petri dishes containing solid MEX medium. The RUT-C30Δzface1-1 shows a higher result, with 1.42-fold greater than the parental strain. The mutants RUT-C30Δzface1-2 and RUT-C30Δzface1-3 presented the enzymatic index, increased by 1.21 and 1.16-fold than RUT-C30 lineage (Figure 5 A).
Hydrolysis tests. (A) Comparison between enzymatic indices in 96 hours of incubation. Petri dishes containing mutant and parental lines were incubated at 28 ° C for 96 hours. After this period, the colonies were stained with Congo red (2.5 g/L), and the diameters of hydrolysis and colony were measured. The division of the diameter of hydrolysis by the diameter of the colony estimated the enzymatic index. Statistical analysis was performed according to Dunnett's test using significance level of p < 0.05, in which (*) p < 0.05 and (**) p < 0.01. (B) Hydrolysis of crushed sugarcane bagasse by mutant strains compared to RUT-C30. Statistical analysis was performed according to Dunnett's test using significance level of p < 0.05, in which (*) p <0.05 and (**) p <0.01. Error bars represent the standard deviation of the results for each variable.
In order to verify the efficiency in the biomass hydrolysis, the enzymatic extracts of the mutants and RUT-C30 strain were incubated with sugarcane bagasse for 24 hours. After that, the reducing sugar released was measured, and the values from mutant strains were compared to the RUT-C30 strain. The mutants, RUT-C30Δzface1-1 and RUT-C30Δzface1-2, presented an increased hydrolysis that was 1.3 times the size of the RUT-C30 strain. The RUT-C30Δzface1-3 was the one that obtained the best result, presenting a hydrolysis value of the biomass that was 1.4 times greater than the hyper-productive RUT-C30 strain (Figure 5 B).
DISCUSSION
Advances in genetic transformation techniques have made important contributions to molecular genetics. Molecular strategies have been developed with the aim to obtain efficient strains in the production of cellulolytic enzymes [2828 Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013 Jan; 31(3):233-9.]. The overexpression of the bgl1 gene in T. reesei RUT-C30, for example, was one of the techniques that significantly increased the production of β-glycosidase and total cellulose [2929 Zhang J, Zhong Y, Zhao X, Wang T. Development of the cellulolytic fungus Trichoderma reesei strain with enhanced ß-glucosidase and filter paper activity using strong artifical cellobiohydrolase 1 promoter. Bioresour Technol. 2010 Dec; 101(24):9815-8.]. In this way, the assays of this work demonstrate an increase of enzymatic secretion by the T. reesei RUT-C30Δzface1 strains with the codifying zinc finger domain of ACE1 deletion when compared with the RUT-C30 parental strain.
The results of lineage growth rate assays show a significant difference between the mycelial sizes of the mutants and the parental strain and corroborate with the work described by Aro and coauthors [1616 Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003 Jan; 69(1):56-65.]. The authors describe that the growth of lineages with the deletion of ACE1 was superior when compared to the growth of the lineages without deletion. Furthermore, Shah and coauthors [3030 Shah S, Nasreen S, Sheikh PA. Cultural and Morphological Characterization of Trichoderma spp. Associated with Green Mold Disease of Pleurotus spp. in Kashmir. Res J Microbiol. 2012 Feb; 7(2):139-44.] characterized several species of this fungus and found that all analyzed strains differed in both the mycelial growth rate and the appearance of the colonies. Hermosa and coauthors [3131 Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, et al. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol. 2000 May; 66(5):1890-8.] also found different degrees of sporulation in relation to the different species of T. reesei.
The T. reesei RUTC-30 strain produces more cellulases when compared to the wild QM6a strain, because it has the truncated CRE transcription factor [3232 Song Y, Wi SG, Kim HM, Bae HJ. Cellulosic bioethanol production from Jerusalem artichoke (Helianthus tuberosus L.) using hydrogen peroxide-acetic acid (HPAC) pretreatment. Bioresour Technol. 2016 Aug; 214:30-6.]. The mutant strains described in this work show the zinc finger motif of the cellulase expression repressor ACE1, deleted in addition to the truncated CRE and observed in the RUTC-30 strain. This deletion leads to an increase in cellulase and pectinase production and suggest a probable action of ACE1 on expression regulation of further glycoside hydrolases beyond cellulases. However, a significative effect on xylanase or invertase production by the mutants was not observed. The cellulase values produced by the mutants demonstrate success in the optimization of the cellulolytic capacity of T. reesei, and, consequently, stand out as an important ally for the increase of bioethanol production. Ivanova and coauthors [3333 Ivanova C, Bååth JA, Seiboth B, Kubicek CP. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PloS One 2013 May; 8(5):1-10.] report that lactose is the only source of soluble carbon that induces T. reesei cellulases at an industrial level. Herpoël-Gimbert and coauthors [3434 Herpoël-Gimbert I, Margeot A, Dolla A, Jan G, Mollé D, Lignon S, et al. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol Biofuels 2008 Dec; 1(1):18-30.] also demonstrated lactose as a source of carbon to improve the yield of cellulases in the RUT-C30 and CL847 lines. According to Aro and coauthors [1616 Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003 Jan; 69(1):56-65.], strains with deleted ace1 are able to degrade cellulose more efficiently. This report corroborates with the results in the present study, in which increased production of cellulases occurred in mutants when compared to RUT-C30. In addition, this result demonstrates that the ace1 gene is linked to the regulation of cellulase production, corroborating with the literature [1515 Castro LS, Antoniêto AC, Pedersoli WR, Silva-Rocha R, Persinoti GF, Silva RN. Expression pattern of cellulolytic and xylanolytic genes regulated by transcriptional factors XYR1 and CRE1 are affected by carbon source in Trichoderma reesei. Gene Expr Patterns. 2014 Mar; 14(2):88-95.,3535 Portnoy T, Margeot A, Siedl-Seiboth V, Le Crom S, Chaabane FB, Linke R, et al Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell. 2011 Feb; 10(2):262-71.].
The mutant strains obtained in this work showed higher hydrolysis index than RUT-C30 strain. In this way, Lopes and coauthors [36Lopes VR, Junior GF, Braga R, Jesus MA, Martins C, Pinto G. [Activity of Xylanase in Strains of Colletrotichum and Trichoderma]. Proceedings of the 17th National Bioprocesses Symposium; 2009; Natal, RN. Natal: Federal University of Rio Grande do Norte (RN); 2009. 7 p. Portuguese.36], using the same technique to determinate enzymatic indices, obtained results of approximately 1.04 and 1.08 for the T666 and T300 strains, respectively. Florencio [2626 Florencio C, Couri S, Farinas CS. Correlation between Agar Plate Screening and Solid-State Fermentation for the Prediction of Cellulase Production by Trichoderma Strains. Enzyme Res. 2012 Nov; 2012:1-7.] evaluated the hydrolytic potential of 78 strains of the genus Trichoderma, belonging to the Embrapa Agroindústria Tropical (Fortaleza, CE), Embrapa Meio Ambiente (Jaguariúna, SP), and Embrapa Recursos Genetics and Biotechnology (Brasília, DF). Florencio [2626 Florencio C, Couri S, Farinas CS. Correlation between Agar Plate Screening and Solid-State Fermentation for the Prediction of Cellulase Production by Trichoderma Strains. Enzyme Res. 2012 Nov; 2012:1-7.] also concluded that the RUT-C30 was a lineage with a greater power of hydrolysis of cellulases. The RUT-C30Δzface1 mutant strains described in this work presented a higher power of hydrolysis than RUT-C30 and, therefore, is a lineage that deserves attention in the search for cellulase optimization by T. reesei.
The hydrolysis of biomass was a crucial result to confirm the creation of strains that could be used in the ethanol production from the lignocellulosic residue. In this assay, it was possible to observe a significant difference between all the mutants, in relation to parental lineage, especially the RUT-C30Δzface1-3 lineage, which showed 1.4 times more biomass hydrolysis than the strain RUT-C30. The conversion of plant-derived carbohydrates into bioethanol is one of the main sources of renewable energy to obtain biofuels. One step for producing this fuel is the hydrolysis of the cell wall polysaccharides by using enzymes, such as cellulases [3737 Souza AP, Leite DCC, Pattathil S, Hahn MG, Buckeridge MS. Composition and structure of sugarcane cell wall polysaccharides: Implications for second-generation bioethanol production. Bioenergy Res. 2013 Nov; 6:564-79.]. The cellulose hydrolysis produces glucose that will be used in the bioethanol production process [3838 Buckeridge MS, de Souza AP, Arundale RA, Anderson-Teixeira KJ, DeLucia E. Ethanol from sugarcane in Brazil: a 'midway' strategy for increasing ethanol production while maximizing environmental benefits. Glob Change Biol Bioenergy. 2012 Sep; 4:119-26.]. In this regard, some strategies are currently being developed to reduce the cost of bioethanol production, such as improving the biological pretreatment of lignocellulosic biomass with optimized strains of cellulase producers [3939 García-Torreiro M, López-Abelairas M, Lu-Chau TA, Lema JM. Fungal pretreatment of agricultural residues for bioethanol production. Ind Crops Prod. 2016 Oct; 89:486-92.]. Thus, the production enhancement of cellulase, due to the genetic modification of the T. reesei RUT-C30 fungal strain, will certainly improve the viability of the bioethanol production.
The RUT-C30 is reported as hyper-productive of cellulase [1313 Peterson R, Nevalainen H. Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology. 2012 Jan; 158(1):58-68.], and the mutants elaborated in this work present both plate hydrolysis and biomass hydrolysis, which are larger than the RUT-C30 strain. Considering that the hydrolysis process can account for 40% of the total value of bioethanol production [1212 Bendig C, Weuster-Botz D. Reaction engineering analysis of cellulase production with Trichoderma reesei RUT-C30 with intermittent substrate supply. Bioprocess Biosyst Eng. 2013 Jul; 36(7):893-900.], RUT-C30Δzface1 mutants are of great importance for the use of biomass as a source to become economically viable on an industrial scale.
CONCLUSION
Cellulose and hemicellulose represent the largest source of renewable energy on earth and could be widely used for biofuel production. Although there are chemical processes for the hydrolysis of these compounds, the cost and low yield of these processes are a major problem for the use of biomass on an industrial scale.
Currently, the RUT-C30 lineage is the most industrially used lineage and, therefore, is the target of research for the creation of strains that have to produce cellulolytic enzymes more efficiently. For this purpose, the RUT-C30Δzface1-1, RUT-C30Δzface1-2, and RUT-C30Δzface1-3 strains were created by partial deletion of the cellulase repressor gene ACE1 in the RUT-C30 strain. The mutants of this work showed greater efficiency in the enzymatic activity and sugarcane hydrolysis when compared to the RUT-C30 hyper-productive strain.
The increase of the enzymatic production by the mutants of this work shows that the strains-now known as cellulase hyperproducers-can have their enzymatic production optimized through genetic engineering. Thus, the T. reesei lines of RUT-C30Δzface1 are promising when applied in biorefineries, in an attempt to leverage the use of sugarcane residue as a renewable energy source, thus, contributing to the establishment of bioethanol as a viable alternative in the fuel market.
Acknowledgments
The authors thank the Coordination for the Improvement of Higher Education Personnel (CAPES), Araucária Foundation, and Paraná Western State University (UNIOESTE).
REFERENCES
-
1Gupta A, Verma JP. Sustainable bio-ethanol production from agro-residues: a review. Renew Sust Energ Rev. 2015 Jan; 41:550-67.
-
2Popp J, Lakner Z, Harangi-Rákos M, Fári M. The effect of bioenergy expansion: food, energy, and environment. Renew Sust Energ Rev. 2014 Apr; 32:559-78.
-
3Raghuwanshi S, Deswal D, Karp M, Kuhad RC. Bioprocessing of enhanced cellulase production from a mutant of Trichoderma asperellum RCK2011 and its application in hydrolysis of cellulose. Fuel (Lond). 2014 May; 124:183-9.
- Santos FA, Queiróz JD, Colodette JL, Fernandes SA, Guimarães VM, Rezende ST. [Potential of sugarcane straw for ethanol production]. Quim Nova. 2012 Jan; 35(5):1004-10. Portuguese.4
-
5Chiaramonti D, Prussi M, Ferrero S, Oriani L, Ottonello P, Torre P, et al. Review of pretreatment processes for lignocellulosic ethanol production, and development of an innovative method. Biomass Bioenerg. 2012 Nov; 46:25-35.
-
6Gomes D, Rodrigues AC, Domingues L, Gama M. Cellulase recycling in biorefineries-is it possible? Appl Microb Biotechnol. 2015 May; 99(10):4131-43.
- Imamoglu E, Sukan FV. Scale-up and kinetic modeling for bioethanol production. Bioresour Technol. 2013 Sept; 144:311-20.7
-
8Ruiz HA, Silva DP, Ruzene DS, Lima LF, Vicente AA, Teixeira JA. Bioethanol production from hydrothermal pretreated wheat straw by a flocculating Saccharomyces cerevisiae strain-Effect of process conditions. Fuel. 2012 May; 95:528-36.
-
9Baeyens J, Kang Q, Appels L, Dewil R, Lv Y, Tan T. Challenges and opportunities in improving the production of bio-ethanol. Prog Energy Combust Sci. 2015 Apr; 47:60-88.
-
10Sarkar N, Ghosh SK, Bannerjee S, Aikat K. Bioethanol production from agricultural wastes: An overview. Renew Energy. 2012 Jan; 37(1):19-27.
-
11Sukumaran RK, Singhania RR, Pandey A. Microbial cellulases-production, applications and challenges.J Sci Ind Res (India). 2005 Nov; 64(11):832-44.
-
12Bendig C, Weuster-Botz D. Reaction engineering analysis of cellulase production with Trichoderma reesei RUT-C30 with intermittent substrate supply. Bioprocess Biosyst Eng. 2013 Jul; 36(7):893-900.
-
13Peterson R, Nevalainen H. Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology. 2012 Jan; 158(1):58-68.
-
14Kubicek CP. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J Biotechnol. 2013 Jan; 163(2):133-42.
-
15Castro LS, Antoniêto AC, Pedersoli WR, Silva-Rocha R, Persinoti GF, Silva RN. Expression pattern of cellulolytic and xylanolytic genes regulated by transcriptional factors XYR1 and CRE1 are affected by carbon source in Trichoderma reesei. Gene Expr Patterns. 2014 Mar; 14(2):88-95.
-
16Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003 Jan; 69(1):56-65.
-
17Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992 Jan; 110(1):119-22.
-
18Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, et al. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels 2012 Jan; 5(1):1-10.
-
19Mota Júnior AO, Malavazi I, Soriani FM, Heinekamp T, Jacobsen I, Brakhage AA, et al. Molecular characterization of the Aspergillus fumigatus NCS-1 homologue, NcsA. Mol Genet Genomics 2008 Dec; 280(6):483-95.
-
20Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006 Jul; 103(27):10352-7.
- Cassago A, Panepucci R, Baião A, Henrique-Silva F. Cellophane based mini-prep method for DNA extraction from the filamentous fungus Trichoderma reesei. BMC Microbiol. 2002 Jun; 2,14-8.21
-
22Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19(1):11-5.
-
23Plaza GA, Upchurch R, Brigmon RL, Whitman WB, Ulfig K. Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Pol J Environ Stud. 2004 Jan; 13(3):315-8.
-
24Ahamed A, Vermette P. Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochem Eng J. 2008 Jul; 40(3):399-407.
-
25Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959 Mar; 31(3):426-8.
-
26Florencio C, Couri S, Farinas CS. Correlation between Agar Plate Screening and Solid-State Fermentation for the Prediction of Cellulase Production by Trichoderma Strains. Enzyme Res. 2012 Nov; 2012:1-7.
-
27Ribeiro LF, de Lucas RC, Vitcosque GL, Ribeiro LF, Ward RJ, Rubio MV, et al. A novel thermostable xylanase GH10 from Malbranchea pulchella expressed in Aspergillus nidulans with potential applications in biotechnology. Biotechnol Biofuels 2014 Jul; 7(1):115-126.
-
28Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013 Jan; 31(3):233-9.
-
29Zhang J, Zhong Y, Zhao X, Wang T. Development of the cellulolytic fungus Trichoderma reesei strain with enhanced ß-glucosidase and filter paper activity using strong artifical cellobiohydrolase 1 promoter. Bioresour Technol. 2010 Dec; 101(24):9815-8.
-
30Shah S, Nasreen S, Sheikh PA. Cultural and Morphological Characterization of Trichoderma spp. Associated with Green Mold Disease of Pleurotus spp. in Kashmir. Res J Microbiol. 2012 Feb; 7(2):139-44.
-
31Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, et al. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol. 2000 May; 66(5):1890-8.
-
32Song Y, Wi SG, Kim HM, Bae HJ. Cellulosic bioethanol production from Jerusalem artichoke (Helianthus tuberosus L.) using hydrogen peroxide-acetic acid (HPAC) pretreatment. Bioresour Technol. 2016 Aug; 214:30-6.
-
33Ivanova C, Bååth JA, Seiboth B, Kubicek CP. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PloS One 2013 May; 8(5):1-10.
-
34Herpoël-Gimbert I, Margeot A, Dolla A, Jan G, Mollé D, Lignon S, et al. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol Biofuels 2008 Dec; 1(1):18-30.
-
35Portnoy T, Margeot A, Siedl-Seiboth V, Le Crom S, Chaabane FB, Linke R, et al Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell. 2011 Feb; 10(2):262-71.
- Lopes VR, Junior GF, Braga R, Jesus MA, Martins C, Pinto G. [Activity of Xylanase in Strains of Colletrotichum and Trichoderma]. Proceedings of the 17th National Bioprocesses Symposium; 2009; Natal, RN. Natal: Federal University of Rio Grande do Norte (RN); 2009. 7 p. Portuguese.36
-
37Souza AP, Leite DCC, Pattathil S, Hahn MG, Buckeridge MS. Composition and structure of sugarcane cell wall polysaccharides: Implications for second-generation bioethanol production. Bioenergy Res. 2013 Nov; 6:564-79.
-
38Buckeridge MS, de Souza AP, Arundale RA, Anderson-Teixeira KJ, DeLucia E. Ethanol from sugarcane in Brazil: a 'midway' strategy for increasing ethanol production while maximizing environmental benefits. Glob Change Biol Bioenergy. 2012 Sep; 4:119-26.
-
39García-Torreiro M, López-Abelairas M, Lu-Chau TA, Lema JM. Fungal pretreatment of agricultural residues for bioethanol production. Ind Crops Prod. 2016 Oct; 89:486-92.
HIGHLIGHTS
-
1
Enhance of Trichoderma reesei RUT-C30∆zface1.
-
2
Deletion of zinc finger of the repressor transcription factor cellulase ACE1.
-
3
Optimized fungal strain for the production of cellulase.
-
4
Greater efficiency in the enzymatic activity and sugarcane hydrolysis.
-
Funding:
This research was funded by National Council for Scientific and Technological Development (CNPq), grant number 458859/2014-1.
Publication Dates
-
Publication in this collection
15 May 2020 -
Date of issue
2020
History
-
Received
27 Mar 2019 -
Accepted
14 Feb 2020