Abstract
Continuous occurrence of heavy metals is a major cause of environmental pollution due to its toxic effects. At minimum concentrations, these metals are highly reactive and can gather in the food chains and food web, causing major dangers to public health concerns. Soil samples were collected from Paharang drain, Faisalabad. Cadmium tolerant bacteria were isolated and evaluated for their MIC against Cd. The isolated bacterial strain GCFSD01 showed MIC value upto 30 mM/L. The bacterial strain with the highest resistance against Cd was selected for further study. Molecular characterization of bacterial isolate GCFSD01 was performed by 16S rRNA which confirmed it as Bacillus cereus. Optimum growth conditions of bacterial strain were also evaluated. Strain GCFSD01 showed optimum growth at pH 7 and 37 °C temperature. Our result revealed that B. cereus strain GCFSD01 reduced 61.3% Cd after 48 hrs. Multiple metal tolerance and Cd reduction by B. cereus indicate its potential for further use for decontamination of polluted soil.
Keywords:
textile effluents; industrial hub; metal contaminated soil; heavy metals; Bacillus cereus; biosorption; biodegradation
Resumo
A ocorrência contínua de metais pesados é uma das principais causas de poluição ambiental devido aos seus efeitos tóxicos. A contaminação por metais pesados representa um grande risco para todas as formas de vida encontradas no meio ambiente. Em concentrações mínimas, esses metais são altamente reativos e podem se acumular nas cadeias alimentares e na cadeia alimentar, causando grandes perigos às preocupações com a saúde pública. Amostras de solo foram coletadas no esgoto de Paharang, Faisalabad. Bactérias tolerantes ao cádmio foram isoladas da amostra coletada pelo método da placa de ágar. As colônias separadas individuais selecionadas foram avaliadas quanto às suas concentrações inibitórias mínimas contra Cd. A cepa bacteriana isolada GCFSD01 apresentou valores de CIM de 30 mM/L. A colônia bacteriana que apresentou maior resistência contra o Cd foi selecionada para identificação. Após seleção da maior colônia bacteriana resistente ao Cd, coloração de Gram e diferentes testes bioquímicos foram realizados para a caracterização da bactéria isolada. A caracterização molecular do isolado bacteriano GCFSD01 foi realizada por PCR 16S rRNA confirmando a presença de Bacillus cereus. Após a identificação molecular, as condições ótimas de crescimento da cepa bacteriana também foram verificadas. A cepa GCFSD01 apresentou crescimento ótimo em pH 7 e temperatura de 37 °C. Nosso resultado revelou que a cepa de B. cereus GCFSD01 reduziu 61,3% de Cd após 48 horas. A tolerância a múltiplos metais e a redução de Cd por B. cereus indicam seu potencial para uso posterior na descontaminação do solo poluído.
Palavras-chave:
efluentes têxteis; cubo industrial; solo contaminado por metais; metais pesados; Bacillus cereus; biossorção; biodegradação
1. Introduction
Metal pollution is one of the most persistent and complex environmental issues, causing threat to the ecosystem and human health (Das et al., 2016DAS, S., DASH, H.R. and CHAKRABORTY, J., 2016. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Applied Microbiology and Biotechnology, vol. 100, no. 7, pp. 2967-2984. http://dx.doi.org/10.1007/s00253-016-7364-4. PMid:26860944.
http://dx.doi.org/10.1007/s00253-016-736...
). The discharge of untreated wastewater containing heavy metals in the ecosystem is one of the most important environmental and health challenges in our society (Igiri et al., 2018IGIRI, B.E., OKODUWA, S.I., IDOKO, G.O., AKABUOGU, E.P., ADEYI, A.O. and EJIOGU, I.K., 2018. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. Journal of Toxicology, vol. 1, no. 1, pp. 1-16. http://dx.doi.org/10.1155/2018/2568038. PMid:30363677.
http://dx.doi.org/10.1155/2018/2568038...
; Phillips and Rainbow, 2013PHILLIPS, D.J. and RAINBOW, P.S., 2013. Biomonitoring of trace aquatic contaminants. Guildford: Springer Science & Business Media, pp. 1-371. vol. 37, no. 1.). Toxic metals are highly reactive even at present in low concentrations. Cadmium, a highly toxic, non-biodegradable and non-essential heavy metal that is harmful even at relatively low concentrations (0.001-0.1 mg/L) to organisms and has been suggested to be human carcinogen (Wu et al., 2016WU, X., COBBINA, S.J., MAO, G., XU, H., ZHANG, Z. and YANG, L., 2016. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environmental Science and Pollution Research International, vol. 23, no. 9, pp. 8244-8259. http://dx.doi.org/10.1007/s11356-016-6333-x. PMid:26965280.
http://dx.doi.org/10.1007/s11356-016-633...
). Prolonged and persistent use of polluted water for irrigation of agricultural lands and as feeding source for animals and humans might have toxic effects (Sharaf et al., 2019SHARAF, S., KHAN, M.U.R., ASLAM, A. and RABBANI, M., 2019. Comparative study of heavy metals residues and histopathological alterations in large ruminants from selected areas around industrial waste drain. Pakistan Veterinary Journal. http://dx.doi.org/10.29261/pakvetj/2019.111.
http://dx.doi.org/10.29261/pakvetj/2019....
). In literature, different methods has been used to remediate Cd pollution like chemical and physical but these approaches are costly and often generate toxic secondary waste products that are also harmful to the soil (Das et al., 2016DAS, S., DASH, H.R. and CHAKRABORTY, J., 2016. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Applied Microbiology and Biotechnology, vol. 100, no. 7, pp. 2967-2984. http://dx.doi.org/10.1007/s00253-016-7364-4. PMid:26860944.
http://dx.doi.org/10.1007/s00253-016-736...
). This situation amphasizes to find out eco-friendly and cost effective methods for remediation of heavy metals (Yasar et al., 2013YASAR, A., KHAN, M., TABINDA, A.B., HAYYAT, M.U. and ZAHEER, A., 2013. Percentage uptake of heavy metals of different macrophytes in stagnant and flowing textile effluent. Journal of Animal and Plant Sciences, vol. 23, no. 1, pp. 1709-1713.). Therefore, among the alternatives, the use of microbes for the removal of heavy metals has gained great attention due to many advantages including cheap production, their ability to treat large area of soil and ecofriendly nature (Congeevaram et al., 2007CONGEEVARAM, S., DHANARANI, S., PARK, J., DEXILIN, M. and THAMARAISELVI, K., 2007. Biosorption of chromium and nickel by heavy metal resistant fungal and bacterial isolates. Journal of Hazardous Materials, vol. 146, no. 1-2, pp. 270-277. http://dx.doi.org/10.1016/j.jhazmat.2006.12.017. PMid:17218056.
http://dx.doi.org/10.1016/j.jhazmat.2006...
).
Microorganisms that are able to remove heavy metals are of great interest as bioremediation agents because they can achieve it due to their high surface to volume ratios (Vijayaraghavan and Yun, 2008VIJAYARAGHAVAN, K. and YUN, Y.S., 2008. Bacterial biosorbents and biosorption. Biotechnology Advances, vol. 26, no. 3, pp. 266-291. http://dx.doi.org/10.1016/j.biotechadv.2008.02.002. PMid:18353595.
http://dx.doi.org/10.1016/j.biotechadv.2...
). Gram-negative bacterial cell wall composed of phospholipids, lipoplysaccharides and polypeptidoglycan of which highly charging nature and anionic character provide metal binding (Silhavy et al., 2010SILHAVY, T.J., KAHNE, D. and WALKER, S., 2010. The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology, vol. 2, no. 5, pp. a000414. http://dx.doi.org/10.1101/cshperspect.a000414. PMid:20452953.
http://dx.doi.org/10.1101/cshperspect.a0...
; Das et al., 2016DAS, S., DASH, H.R. and CHAKRABORTY, J., 2016. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Applied Microbiology and Biotechnology, vol. 100, no. 7, pp. 2967-2984. http://dx.doi.org/10.1007/s00253-016-7364-4. PMid:26860944.
http://dx.doi.org/10.1007/s00253-016-736...
). Some bacteria having above can also detoxify heavy metals and produce chelating agents that can bind metal ions while some microorganisms have the ability to transform toxic forms into less toxic forms. Normally indigenous microorganisms adopt themselves under changing environmental conditions and could be a better option to cope these changes (Ndi and Barton, 2012NDI, O.L. and BARTON, M.D., 2012. Resistance determinants of Pseudomonas species from aquaculture in Australia. Journal of Aquaculture Research & Development, vol. 3, no. 1, pp. 1-6. http://dx.doi.org/10.4172/2155-9546.1000119.
http://dx.doi.org/10.4172/2155-9546.1000...
).
Faisalabad being the 3rd biggest city of Pakistan is the industrial hub and known as “Manchester of Pakistan” (Jaffrelot, 2002JAFFRELOT, C., 2002. Nationalism without a nation. 1st ed. Pakistan: In Zed Books.). Heavy industries especially textile industry producing different types of waste are directly discharging their waste into Paharang drain passes through Faisalabad. Therefore, the aim of present study was to isolate, identify and characterize heavy metal tolerant indigenous bacteria and to investigate their biosorptive potential against heavy metals.
2. Material and Methods
2.1. Study area and sample collection
Samples were collected from Faisalabad area (31°25’0”N/73°5’0”E). Industrial effluents are major source of heavy metal contamination in this area. Industries are directly discharging these wastes in to river Chenab through Paharang drain that originates from Chak Jhumra and passes through Faisalabad with collection of these effluents from industrial hub areas. Samples of soil were collected in pre-cleaned plastic bottles. Samples were collected from 0-10 cm depth of ground surface from approximately 500 meters distance intervals. Total 25 soil samples were collected in triplicate.
2.2. Heavy metals analysis of collected samples
After collection, heavy metals were determined in the samples using the method previously described by Zeiner et al. (2007)ZEINER, M., REZIĆ, I. and STEFFAN, I., 2007. Analytical methods for the determination of heavy metals in the textile industry. Kemija u Industriji, vol. 56, no. 11, pp. 587-595.. Cadmium (Cd), cobalt (Co), nickel (Ni), chromium (Cr), zinc (Zn) and lead (Pb) were included for analysis. Samples were digested and metal analysis was performed using Atomic Absorption Spectrophotometer (AAS) (Zeeman Atomic Absorption Spectrophotometer, ZA3000 Series). On the basis of the results, cadmium (Cd) was selected for further study.
2.3. Selection and isolation of cadmium resistant bacteria
Soil samples were taken in sterile water and to isolate bacteria from soil, samples were shaken at 150 rpm for 2 hr. After that, samples were settled for 5 min and one mL of the suspension was serially diluted upto 10−6. Each dilution was spread onto an LB plate supplemented with 1mM Cd+2 and incubated at 37 °C. The growing colonies were observed and again inoculated onto LB plates containing Cd to get pure culture.
2.4. Determination of Minimum Inhibitory Concentration (MIC)
To evaluate the minimum inhibitory concentration (MIC) of the isolated bacteria, MS agar (Minimal Salt agar), (0.1% K2HPO4, 0.1% MgSO4, 1% (NH4)2SO4, 0.02% FeSO4, 0.086% CaCl2 and 0.05% Glucose) was prepared supplemented with Cd ranging from 1mM Cd+2 to 30mM. Each isolate was inoculated on these plates and incubated at 37 °C for 24h. The minimum Cd+2 concentrations at which bacterial isolate did not show growth was referred as its MIC.
2.5. Identification of bacterial isolate
After selecting Cd resistant bacterial isolate, identification was done on the basis of cultural characteristics, gram’s staining and biochemical characteristics like oxidase reaction, catalase reaction, starch hydrolysis, fermentation tests and glucose utilization.
2.6. 16S ribosomal RNA (rRNA) sequencing
For the molecular characterization of bacterial isolate, two primers 27F 5-AGAGTTTGATCCTGGCTCAG-3 and 1522R 5-AAGGAGGTGTGCCARCCGCA-3 were used (Balashov and Humayun 2003BALASHOV, S. and HUMAYUN, M.Z., 2003. Escherichia coli cells bearing a ribosomal ambiguity mutation in rpsD have a mutator phenotype that correlates with increased mistranslation. Journal of Bacteriology, vol. 185, no. 16, pp. 5015-5018. http://dx.doi.org/10.1128/JB.185.16.5015-5018.2003. PMid:12897024.
http://dx.doi.org/10.1128/JB.185.16.5015...
). Total genomic DNA was extracted by CTAB method (Wilson, 2001WILSON, K., 2001. Preparation of genomic DNA from bacteria. Current Protocols in Molecular Biology, vol. 56, no. 15, pp. 2-4. https://doi.org/10.1002/0471142727.mb0204s56.
https://doi.org/10.1002/0471142727.mb020...
). To attain a final volume of 25 mL, a reaction mixture contained template DNA, nuclease-free water, 12.5 mL 2X PCR master mix (Ferments) and primers was prepared. PCR was carried out with the help of thermocycler (Advance Primus 96) by using initial denaturation at 94 °C for 3 min, 35 cycles of denaturation at 94 °C for 45 sec, primer annealing at 50 °C for 60 sec, DNA extension at 72 °C for 90 sec and final extension at 72 °C for 10 min.
For ribotyping, PCR product was sent for commercial sequencing to Macrogen (Korea). The sequences obtained were checked for base calling using FinchTV and contigs made using NCBI BLAST (two sequence alignments). After amplification, 16S rRNA sequence was aligned using ClustalW and compared with known sequences in the GenBank database.
2.7. Determination of optimum growth conditions for GCFSD01 strain
Optimum growth conditions of bacterial isolate were determined with respect to pH and temperature. Bacterial isolate was grown in LB broth at different incubating temperatures viz, 20, 30, 37, 45 and 55 °C for the determination of optimum temperature in the presence and absence of Cd. Their absorbance was determined at 600 nm after 24 h. For pH optimization, pH of the LB broth was adjusted at 5, 6, 7, 8, 9 and 10. OD was measured at 600 nm after 24 h incubation.
2.8. Effect of temperature and pH on Cd2+ uptake ability of bacterial isolate
Effect of pH and temperature on the Cd2+ uptake ability of the bacterial isolate was determined. Bacteria was grown in LB broth supplemented with 1mM Cd2+ at different incubating temperatures ranging from 25 °C to 42 °C and pH ranging from 5 to 9. After 24h of incubation, samples were collected and centrifuged for 5 min at 3000 rpm. Supernatants and pellets were used to evaluate the quantity of Cd2+ by atomic absorption spectrophotometer (Abbas et al., 2014ABBAS, S.Z., RAFATULLAH, M., ISMAIL, N. and LALUNG, J., 2014. Isolation, identification, and characterization of cadmium resistant Pseudomonas sp. M3 from industrial wastewater. Journal of Waste Management, vol. 2014, pp. 160398. http://dx.doi.org/10.1155/2014/160398.
http://dx.doi.org/10.1155/2014/160398...
).
2.9. Multi Metal Resistance (MMR) and antibiotic resistance
Multi metal resistance of isolated strain was determined as described by (Saini and Pant, 2016SAINI, S. and PANT, S., 2016. Heavy metal and antibiotic resistant bacteria isolated from soil contaminated by the sugar industry effluent. International Journal of Biosciences, vol. 8, no. 1, pp. 36-46.). LB agar medium was prepared, autoclaved and filtered sterilized solution of Cd, Cr, Pb and Ni in equal concentration i.e. 1:1:1:1 incorporated in the medium. Antibiotic susceptibility testing of isolated strain GCFSD01 was carried out using disc diffusion method. Antibiotic discs used were: ceftazidime (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), Amikacin (30 μg), piperacillin-tazobactam (100/10 μg), meropenem (10 μg), erythromycin (15 μg) chloramphenicol (30 μg), streptomycin (10 μg), ampicilline (10 μg), rifampicine (5 μg), kanamycin (30 μg) and tetracyclin (30 μg). The interpretation of susceptibility results was done according to the guidelines of Clinical Laboratory Standards Institute (CLSI, 2016CLINICAL AND LABORATORY STANDARDS INSTITUTE – CLSI, 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Wayne: CLSI. http://dx.doi.org/10.1086/510431.
http://dx.doi.org/10.1086/510431...
).
2.10. Evaluation of Cd reduction potential
Reduction potential of strain GCFSD01 (LT985360) was determined against Cd. For this, LB broth was prepared supplemented with 100 mg/L of Cd. After autoclave one flask was inoculated with 2 mL of 24h bacterial culture having turbidity equal to 0.5 McFarland. Flasks were incubated on a shaker at 28 °C for 24, 48 and 72 hr then centrifuged at 8,000 rpm for 15 min and supernatants were collected and Cd concentration was determined through atomic absorption spectrophotometer (Kumar et al., 2010KUMAR, A., BISHT, B.S. and JOSHI, V.D., 2010. Biosorption of heavy metals by four acclimated microbial species, Bacillus spp., Pseudomonas spp., Staphylococcus spp. and Aspergillus niger. Journal of Biological and Environmental Sciences, vol. 4, no. 12, pp. 97-108.).
2.11. Fourier Transform Infrared Spectroscopy (FTIR)
Isolate cultured in presence and absence of Cd were harvested by centrifugation at 4000×G for 10 minutes and washed three times with 1xPBS (phosphate buffer saline). The cell pellets were placed in bench top freeze drier (Labconco™, Missouri, United States of America) overnight. The dried cell pellets were used for infrared spectroscopic analysis and spectra was recorded on FTIR spectrometer (Bruker, Massachusetts, United States of America). The FTIR spectrum was recorded in the region 4000 to 500 cm-1 wave number (Khan, 2013KHAN, S.J., 2013. Effect of HRT on SBR performance for treatability of combined domestic and textile wastewaters. Journal of Chemosphere Society in Pakistan, vol. 35, no. 2, pp. 527-532.).
3. Results
3.1. Heavy metals analysis in collected soil samples
Heavy metal analysis of soil samples through atomic absorption spectrophotometer revealed the presence of different heavy metals in the samples. Concentrations of different heavy metals are: Zn 121.7 ± 0.30, Cr 80.3 ± 0.13, Pb 57. ± 0.07, Cd 11.0 ± 0.03, Cu 36.8 ± 0.04 and Ni 36.5 ± 0.09. Due to higher concentration of Cd in soil, it was selected for further study.
3.2. Isolation of Cd resistant bacterial species from soil samples
Out of 25 samples, only 15 samples exhibited positive growth on medium containing Cd. 15 isolated bacteria using spread plate procedure labeled as GCFSD-01 to GCFSD-15 showed resistance to cadmium were selected for minimum inhibitory concentrations (MIC).
3.3. Determination of minimum inhibitory concentrations
MIC of selected isolates was determined. All selected bacterial isolates showed growth up to 10 mM of Cd+2. However, only five isolates, GCFSD-01, GCFSD-05, GCFSD-08, GCFSD-09 and GCFSD-13 showed growth upto 20 mM of Cd+2 while only one isolate GCFSD-01 showed growth at at 30 mM concentrations. From MIC results, GCFSD-01 isolate has been selected for further studies.
3.4. Identification of bacterial isolates GCFSD01
Initial identification up to genus level was done by using standard microbiological techniques. Results revealed G+ve, non-motile rods (as shown in Table 1).
3.5. 16S ribosomal RNA (rRNA) sequencing
Molecular characterization through ribotyping confirmed bacterial strain GCFSD01 as Bacillus cereus and submitted to GenBank with accession number LT98536. The strain displayed highest level of identity 98.44% (query coverage 97%) with B. cereus MB401 (Accession number KJ833772) isolated from food products of Punjab, Pakistan. Second closest relative of our isolate was found B. cereus AHBR1 (KF241514) isolated from household dust samples from Singapore. Finally, evolutionary status of our isolate was confirmed by constructing phylogenetic tree using maximum likelihood method (See Figure 1).
16S rRNA sequence-based phylogenetic tree of Bacillus cereus isolated from Cd contaminated soil.
3.6. Determination of optimum growth conditions for GCFSD01 strain
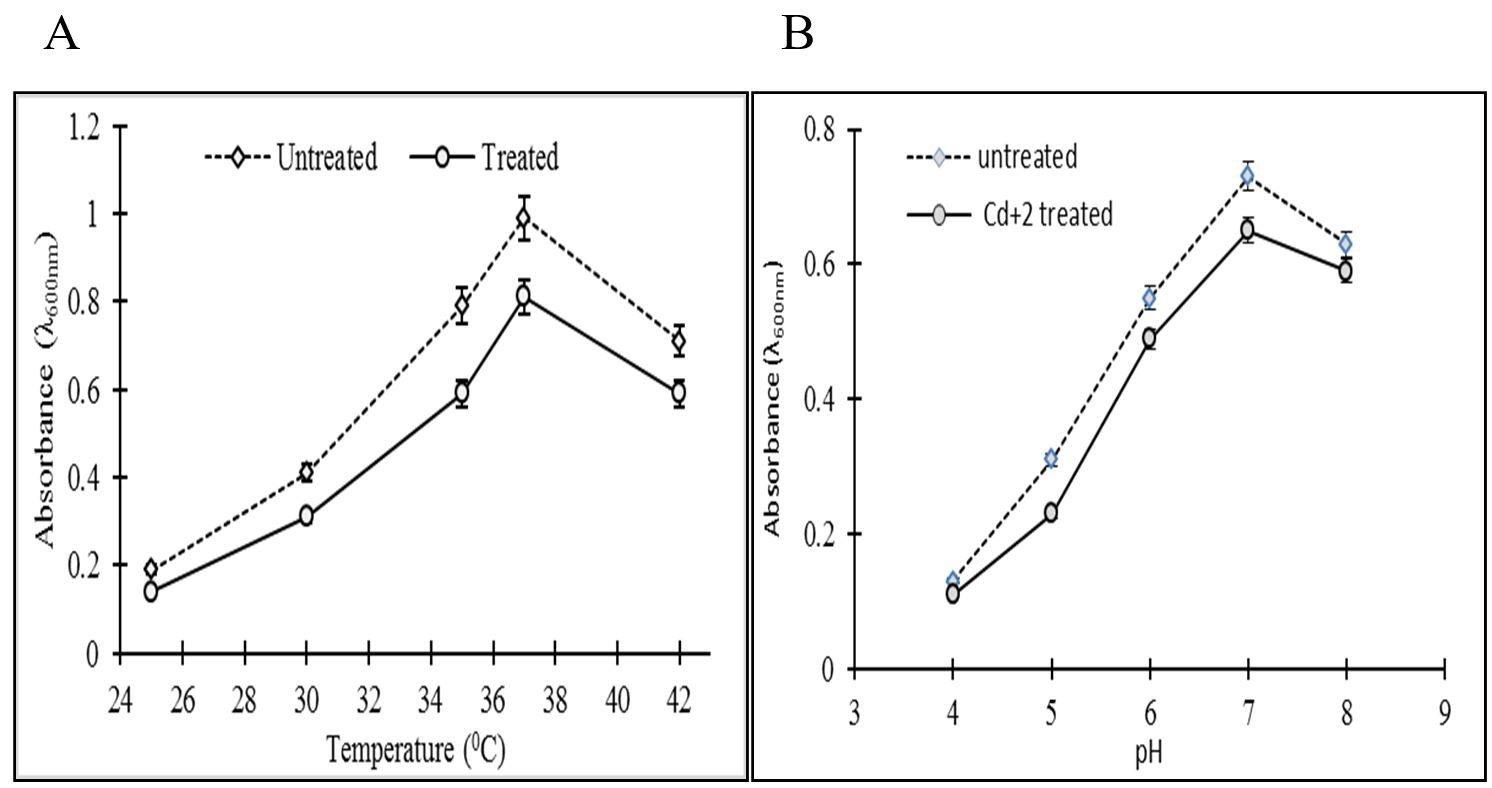
Bacillus cereus GCFSD01 was found to be grown best at 37 °C and optimum pH for its growth was found to be 7 (as shown in Figure 22B).
Graph showing effect of Cd vs. control on growth rate of Bacillus cereus strain GCFSD01: (A) at different temperature; (B) at different pH.
3.7. Effect of temperature and pH on Cd2+ uptake ability of Bacillus cereus GCFSD01
B. cereus GCFSD01 showed variations in its Cd+2 uptake ability with the changes in pH and temperature. At 25 °C B. cereus GCFSD01 was able to remove 50% Cd+2 from the medium. At 37 °C, B. cereus exhibited maximum uptake ability of 77%. While at 42 °C, it removed only 40% Cd+2 form the medium. At pH 5, B. cereus was able to remove 40% Cd+2 from the medium. With the increased pH, Cd+2 uptakes ability of B. cereus also increased. At pH 6, it was able to remove 75% Cd+2 from the medium while further increase in pH caused a decrease in removing ability of Cd+2. At pH 7, it removed 68% Cd+2 while at pH 8, it was able to remove 59% Cd+2 from the medium. At pH 9, it removed only 35% Cd+2 from the medium, respectively.
3.8. Multi Metal Resistance (MMR) and antibiotic resistance
The result of multi metal resistance is presented in Table 2. B. cereus strain GCFSD01 was found to be sensitive to amikacin (30 μg), chloramphenicol (30 μg), kanamycin (30 μg), gentamicin (10 μg), tetracycline (30 μg), ampicillin (10 μg) (as shown in Table 2).
Multi Metal Resistance (MMR) against heavy metals and antibiotics sensitivity profiling of strain GCFSD01.
3.9. Evaluation of Cd reduction potential
Reduction of Cd by of B. cereus GCFSD01 was evaluated using atomic absorption spectrophotometer (AAS). For this, percentage reduction was measured after 24, 48 h and 72 hrs. Our data suggested that strain GCFSD01 reduced 40.2%, 61.3% and 57.6% of Cd (100 mg/L) in 24, 48 h and 72 hrs respectively (as shown in Figure 3).
3.10. FTIR spectrum
The shifts in IR spectrum peaks were noticed upon binding of Cd. Furthermore, attachment of Cd significantly decreased the transmittance (%) and position of multiple peaks. According to our results, carboxyl, amino and hydroxyl groups played important role in binding of Cd by electrostatic interactions. Spectra indicated contribution of amide group (1362.50 cm-1 and 1635.83 cm-1) and hydroxyl group (3261.87 cm-1) in Cd attachment. After binding of Cd, 1635.83 cm-1 shifted to 1635.84 cm-1, 2125.23 cm-1 to 2120.084 cm-1, 2360.04 cm-1 to 2359.98 cm-1, 3261.87 cm-1 to 3263.81 cm-1 due to NH2 scissoring, amide vibration and OH stretching, respectively (as shown in Figure 4).
4. Discussion
Applications of heavy metals resistant bacterial isolates play an important roMle in the reduction of toxic metals from the environment. Effluents discharged from industries harbor bacterial growth by adopted heavy metals resistant mechanisms which making them more resistant to high concentrations of these toxic metals. Modifications in bacterial structures presented their adaptations in high concentrations of heavy metals (Ghani et al., 2012GHANI, A., SAEED, S., ALI, Z., AHMAD, I. and ISHTIAQ, M., 2012. Heavy metals and nutritional composition of some selected herbal plants of Soon Valley, Khushab, Punjab, Pakistan. African Journal of Biotechnology, vol. 11, no. 76, pp. 14064-14068.). Heavy metal-resistant bacterial strains shared common resistant adaptations among them due to the same toxic mechanisms of heavy metals (Ianeva, 2009IANEVA, O.D., 2009. Mechanisms of bacteria resistance to heavy metals. Mikrobiolohichnyi Zhurnal, vol. 71, no. 6, pp. 54-65. PMid:20455433.). Contamination due to high concentrations of heavy metals in the environment posing a serious threat to all life forms living either in aquatic bodies and on land.
In the present study, Cd+2 resistant bacteria was isolated from the soil contaminated with high concentrations of toxic metals from Paharnag main drain, Bawa chak, Faisalabad, Pakistan. It has been reported that 0.001 to 0.21 mg/L of Cd present in ground water samples in different areas of Pakistan. Highest value of 0.21 mg/L was reported in Khyber Pakhtunkhwa (KPK) province industrial area (Waseem et al., 2014WASEEM, A., ARSHAD, J., IQBAL, F., SAJJAD, A., MEHMOOD, M. and MURTAZA, G., 2014. Pollution status of Pakistan: a retrospective review on heavy metal contamination of water, soil, and vegetables. BioMed Research International, vol. 2014, pp. 1-29. http://dx.doi.org/10.1155/2014/813206. PMid:25276818.
http://dx.doi.org/10.1155/2014/813206...
). Cd+2 ions removal by the applications of bacterial biomass has been reported in many studies (Vijayaraghavan and Yun, 2008VIJAYARAGHAVAN, K. and YUN, Y.S., 2008. Bacterial biosorbents and biosorption. Biotechnology Advances, vol. 26, no. 3, pp. 266-291. http://dx.doi.org/10.1016/j.biotechadv.2008.02.002. PMid:18353595.
http://dx.doi.org/10.1016/j.biotechadv.2...
; He et al., 2011HE, M., LI, X., LIU, H., MILLER, S.J., WANG, G. and RENSING, C., 2011. Characterization and genomic analysis of a highly chromate resistant and reducing bacterial strain Lysinibacillus fusiformis ZC1. Journal of Hazardous Materials, vol. 185, no. 2-3, pp. 682-688. http://dx.doi.org/10.1016/j.jhazmat.2010.09.072. PMid:20952126.
http://dx.doi.org/10.1016/j.jhazmat.2010...
; Huang et al., 2014HUANG, F., GUO, C.L., LU, G.N., YI, X.Y., ZHU, L.D. and DANG, Z., 2014. Bioaccumulation characterization of cadmium by growing Bacillus cereus RC-1 and its mechanism. Chemosphere, vol. 109, pp. 134-142. http://dx.doi.org/10.1016/j.chemosphere.2014.01.066. PMid:24560281.
http://dx.doi.org/10.1016/j.chemosphere....
). Bacterial strain has high capacity for the reduction of Cd+2 from the water polluted with toxic metals and has earned great deal in the field of bioremediation. After initial screening, it was observed that 15 out of total 25 soil samples exhibited bacterial growth on LB agar containing 1mM of Cd. MIC of selected bacterial strains were determined and one strain GCFSD01 was able to grow on minimal salt agar incorporated with 30mM of Cd. It was then screened for MMR against Cd, Cr and Pb. Results showed the tolerance of bacteria to Cd upto 30mM and MMR (Cd: Cr: Pb: Ni, 1:1:1:1) up to 5.5 mM. Molecular characterization confirmed the strain as Bacillus sp/ showing 98.44% similarity with B. cereus MB401 (KJ833772). Presented results in this study are accordance to Shamim and Rehman (2012)SHAMIM, S. and REHMAN, A., 2012. Cadmium resistance and accumulation potential of klebsilla pneumonia strain CBL-1 isolated from industrial wastewater. Pakistan Journal of Zoology, vol. 44, no. 1, pp. 203-208. who performed the similar work and isolated the Klebsiella pneumoniae strain CBL-1 and found the Cd tolerance upto 550 ug/mL. Heavy metal tolerant bacteria have been isolated from tannery effluents and found that Bacillus spp exhibited tolerance in this order Pb > Cu and Zn > Cr > Hg. They found the Cd removal upto 93% at 96 hrs but in this study we found the maximum capacity of Cd removal at 48 hrs and after that removal % declined. Pagananelli et al., 2010PAGNANELLI, F., VIGGI, C. C., and TORO, L. 2010. Isolation and quantification of cadmium removal mechanisms in batch reactors inoculated by sulphate reducing bacteria: biosorption versus bioprecipitation.Bioresource technology, vol. 101, no. 9, pp. 2981-2987., reported the difference between Cd removal by bioprecipitation (23%) and biosorption (77%) by the use of sulphate reducing bacteria, thus concluding biosorption method more efficient. Kao et al. (2009)KAO, W.C., WU, J.Y., CHANG, C.C. and CHANG, J.S., 2009. Cadmium biosorption by polyvinyl alcohol immobilized recombinant Escherichia coli. Journal of Hazardous Materials, vol. 169, no. 1-3, pp. 651-658. http://dx.doi.org/10.1016/j.jhazmat.2009.03.140. PMid:19398152.
http://dx.doi.org/10.1016/j.jhazmat.2009...
reported 82.7% Cd removal by E. coli cells with 10 mg/L initial Cd concentration. Shamim and Rehman (2012)SHAMIM, S. and REHMAN, A., 2012. Cadmium resistance and accumulation potential of klebsilla pneumonia strain CBL-1 isolated from industrial wastewater. Pakistan Journal of Zoology, vol. 44, no. 1, pp. 203-208. found that K. pneumoniae capable of decreasing 46%, 68%, 83% and 93% of Cd (100µg/mL) from the medium after 24, 48, 72 and 96 hrs. Our data about Cd removal clearly related with bacterial growth as maximum Cd removal was observed after 48 hrs (61.3% reduction) that is maximum growth phase of our strain as predicted by optimum growth condition of the strain GCFSD01. After that percentage reduction of Cd decreases as shown in our data. Some scientists believed that bacterial growth decreases after maximum bioaccumulation of metal ions (Abbas et al., 2014ABBAS, S.Z., RAFATULLAH, M., ISMAIL, N. and LALUNG, J., 2014. Isolation, identification, and characterization of cadmium resistant Pseudomonas sp. M3 from industrial wastewater. Journal of Waste Management, vol. 2014, pp. 160398. http://dx.doi.org/10.1155/2014/160398.
http://dx.doi.org/10.1155/2014/160398...
).
FTIR analysis was performed to observe the difference between functional groups present in bacterial cell wall in relation to biosorption of Cd. Results revealed the presence of a number of absorption peaks in control as compared to sample with metal stress. It was observed that there was a change in peak at 3261 and 2125 cm−1 showing the involvement of amino and hydroxyl group in Cd biosorption. Afzal et al. (2017)AFZAL, A.M., RASOOL, M.H., WASEEM, M. and ASLAM, B., 2017. Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express, vol. 7, no. 1, pp. 184. http://dx.doi.org/10.1186/s13568-017-0482-2. PMid:28963704.
http://dx.doi.org/10.1186/s13568-017-048...
also reported amino and hydroxyl group role for the Ni and Co absorption. Antibiotic susceptibility testing of B. cereus GCFSD01 against different antibiotics has been exhibited. The results present in this study are in accordance with the Abbas et al. (2017)ABBAS, M., PAUL, M. and HUTTNER, A., 2017. New and improved: a review of novel antibiotics for Gram-positive bacteria. Clinical Microbiology and Infection, vol. 23, no. 10, pp. 697-703. http://dx.doi.org/10.1016/j.cmi.2017.06.010. PMid:28642145.
http://dx.doi.org/10.1016/j.cmi.2017.06....
who demonstrated that Cd resistant Bacillus also harbored the antibiotic resistance. In literature, correlation between metal resistance and antibiotic resistance has been reported. This is due to the result of contaminated environments causing coincidental selection of resistance factors both for heavy metal and antibiotics (Chellaiah, 2018CHELLAIAH, E.R., 2018. Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: a minireview. Applied Water Science, vol. 8, no. 6, pp. 154. http://dx.doi.org/10.1007/s13201-018-0796-5.
http://dx.doi.org/10.1007/s13201-018-079...
). Similar observations on metal and antibiotics resistance have been reported in different bacterial strains (Thacker et al., 2007THACKER, U., PARIKH, R., SHOUCHE, Y. and MADAMWAR, D., 2007. Reduction of chromate by cell-free extract of Brucellasp. isolated from Cr(VI) contaminated sites. Bioresource Technology, vol. 98, no. 8, pp. 1541-1547. http://dx.doi.org/10.1016/j.biortech.2006.06.011. PMid:16931000.
http://dx.doi.org/10.1016/j.biortech.200...
). To detoxify the heavy metals effects, bioremediation approach has gained much attention due to environment friendly, safe and cost effective nature.
On the basis of this study, it was concluded that native bacterial strain GCFSD01 B. cereus isolated from industrial soil of Faisalabad Pakistan possessed Cd tolerance ability and biosorptive potential. However, exact resistance mechanisms needs further investigation but, it may be a potential candidate as bioremediation agents development in future to detoxify contaminated soil at industrial surroundings within natural environments in Pakistan.
Acknowledgements
The facilities and support provided by the Department of Zoology, Government College University Faisalabad are highly acknowledged.
-
(With 4 figures)
References
- ABBAS, M., PAUL, M. and HUTTNER, A., 2017. New and improved: a review of novel antibiotics for Gram-positive bacteria. Clinical Microbiology and Infection, vol. 23, no. 10, pp. 697-703. http://dx.doi.org/10.1016/j.cmi.2017.06.010 PMid:28642145.
» http://dx.doi.org/10.1016/j.cmi.2017.06.010 - ABBAS, S.Z., RAFATULLAH, M., ISMAIL, N. and LALUNG, J., 2014. Isolation, identification, and characterization of cadmium resistant Pseudomonas sp. M3 from industrial wastewater. Journal of Waste Management, vol. 2014, pp. 160398. http://dx.doi.org/10.1155/2014/160398
» http://dx.doi.org/10.1155/2014/160398 - AFZAL, A.M., RASOOL, M.H., WASEEM, M. and ASLAM, B., 2017. Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express, vol. 7, no. 1, pp. 184. http://dx.doi.org/10.1186/s13568-017-0482-2 PMid:28963704.
» http://dx.doi.org/10.1186/s13568-017-0482-2 - BALASHOV, S. and HUMAYUN, M.Z., 2003. Escherichia coli cells bearing a ribosomal ambiguity mutation in rpsD have a mutator phenotype that correlates with increased mistranslation. Journal of Bacteriology, vol. 185, no. 16, pp. 5015-5018. http://dx.doi.org/10.1128/JB.185.16.5015-5018.2003 PMid:12897024.
» http://dx.doi.org/10.1128/JB.185.16.5015-5018.2003 - CHELLAIAH, E.R., 2018. Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: a minireview. Applied Water Science, vol. 8, no. 6, pp. 154. http://dx.doi.org/10.1007/s13201-018-0796-5
» http://dx.doi.org/10.1007/s13201-018-0796-5 - CLINICAL AND LABORATORY STANDARDS INSTITUTE – CLSI, 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria Wayne: CLSI. http://dx.doi.org/10.1086/510431
» http://dx.doi.org/10.1086/510431 - CONGEEVARAM, S., DHANARANI, S., PARK, J., DEXILIN, M. and THAMARAISELVI, K., 2007. Biosorption of chromium and nickel by heavy metal resistant fungal and bacterial isolates. Journal of Hazardous Materials, vol. 146, no. 1-2, pp. 270-277. http://dx.doi.org/10.1016/j.jhazmat.2006.12.017 PMid:17218056.
» http://dx.doi.org/10.1016/j.jhazmat.2006.12.017 - DAS, S., DASH, H.R. and CHAKRABORTY, J., 2016. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Applied Microbiology and Biotechnology, vol. 100, no. 7, pp. 2967-2984. http://dx.doi.org/10.1007/s00253-016-7364-4 PMid:26860944.
» http://dx.doi.org/10.1007/s00253-016-7364-4 - GHANI, A., SAEED, S., ALI, Z., AHMAD, I. and ISHTIAQ, M., 2012. Heavy metals and nutritional composition of some selected herbal plants of Soon Valley, Khushab, Punjab, Pakistan. African Journal of Biotechnology, vol. 11, no. 76, pp. 14064-14068.
- HE, M., LI, X., LIU, H., MILLER, S.J., WANG, G. and RENSING, C., 2011. Characterization and genomic analysis of a highly chromate resistant and reducing bacterial strain Lysinibacillus fusiformis ZC1. Journal of Hazardous Materials, vol. 185, no. 2-3, pp. 682-688. http://dx.doi.org/10.1016/j.jhazmat.2010.09.072 PMid:20952126.
» http://dx.doi.org/10.1016/j.jhazmat.2010.09.072 - HUANG, F., GUO, C.L., LU, G.N., YI, X.Y., ZHU, L.D. and DANG, Z., 2014. Bioaccumulation characterization of cadmium by growing Bacillus cereus RC-1 and its mechanism. Chemosphere, vol. 109, pp. 134-142. http://dx.doi.org/10.1016/j.chemosphere.2014.01.066 PMid:24560281.
» http://dx.doi.org/10.1016/j.chemosphere.2014.01.066 - IANEVA, O.D., 2009. Mechanisms of bacteria resistance to heavy metals. Mikrobiolohichnyi Zhurnal, vol. 71, no. 6, pp. 54-65. PMid:20455433.
- IGIRI, B.E., OKODUWA, S.I., IDOKO, G.O., AKABUOGU, E.P., ADEYI, A.O. and EJIOGU, I.K., 2018. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. Journal of Toxicology, vol. 1, no. 1, pp. 1-16. http://dx.doi.org/10.1155/2018/2568038 PMid:30363677.
» http://dx.doi.org/10.1155/2018/2568038 - JAFFRELOT, C., 2002. Nationalism without a nation 1st ed. Pakistan: In Zed Books.
- KAO, W.C., WU, J.Y., CHANG, C.C. and CHANG, J.S., 2009. Cadmium biosorption by polyvinyl alcohol immobilized recombinant Escherichia coli. Journal of Hazardous Materials, vol. 169, no. 1-3, pp. 651-658. http://dx.doi.org/10.1016/j.jhazmat.2009.03.140 PMid:19398152.
» http://dx.doi.org/10.1016/j.jhazmat.2009.03.140 - KHAN, S.J., 2013. Effect of HRT on SBR performance for treatability of combined domestic and textile wastewaters. Journal of Chemosphere Society in Pakistan, vol. 35, no. 2, pp. 527-532.
- KUMAR, A., BISHT, B.S. and JOSHI, V.D., 2010. Biosorption of heavy metals by four acclimated microbial species, Bacillus spp., Pseudomonas spp., Staphylococcus spp. and Aspergillus niger Journal of Biological and Environmental Sciences, vol. 4, no. 12, pp. 97-108.
- NDI, O.L. and BARTON, M.D., 2012. Resistance determinants of Pseudomonas species from aquaculture in Australia. Journal of Aquaculture Research & Development, vol. 3, no. 1, pp. 1-6. http://dx.doi.org/10.4172/2155-9546.1000119
» http://dx.doi.org/10.4172/2155-9546.1000119 - PAGNANELLI, F., VIGGI, C. C., and TORO, L. 2010. Isolation and quantification of cadmium removal mechanisms in batch reactors inoculated by sulphate reducing bacteria: biosorption versus bioprecipitation.Bioresource technology, vol. 101, no. 9, pp. 2981-2987.
- PHILLIPS, D.J. and RAINBOW, P.S., 2013. Biomonitoring of trace aquatic contaminants Guildford: Springer Science & Business Media, pp. 1-371. vol. 37, no. 1.
- SAINI, S. and PANT, S., 2016. Heavy metal and antibiotic resistant bacteria isolated from soil contaminated by the sugar industry effluent. International Journal of Biosciences, vol. 8, no. 1, pp. 36-46.
- SHAMIM, S. and REHMAN, A., 2012. Cadmium resistance and accumulation potential of klebsilla pneumonia strain CBL-1 isolated from industrial wastewater. Pakistan Journal of Zoology, vol. 44, no. 1, pp. 203-208.
- SHARAF, S., KHAN, M.U.R., ASLAM, A. and RABBANI, M., 2019. Comparative study of heavy metals residues and histopathological alterations in large ruminants from selected areas around industrial waste drain. Pakistan Veterinary Journal http://dx.doi.org/10.29261/pakvetj/2019.111
» http://dx.doi.org/10.29261/pakvetj/2019.111 - SILHAVY, T.J., KAHNE, D. and WALKER, S., 2010. The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology, vol. 2, no. 5, pp. a000414. http://dx.doi.org/10.1101/cshperspect.a000414 PMid:20452953.
» http://dx.doi.org/10.1101/cshperspect.a000414 - THACKER, U., PARIKH, R., SHOUCHE, Y. and MADAMWAR, D., 2007. Reduction of chromate by cell-free extract of Brucellasp. isolated from Cr(VI) contaminated sites. Bioresource Technology, vol. 98, no. 8, pp. 1541-1547. http://dx.doi.org/10.1016/j.biortech.2006.06.011 PMid:16931000.
» http://dx.doi.org/10.1016/j.biortech.2006.06.011 - VIJAYARAGHAVAN, K. and YUN, Y.S., 2008. Bacterial biosorbents and biosorption. Biotechnology Advances, vol. 26, no. 3, pp. 266-291. http://dx.doi.org/10.1016/j.biotechadv.2008.02.002 PMid:18353595.
» http://dx.doi.org/10.1016/j.biotechadv.2008.02.002 - WASEEM, A., ARSHAD, J., IQBAL, F., SAJJAD, A., MEHMOOD, M. and MURTAZA, G., 2014. Pollution status of Pakistan: a retrospective review on heavy metal contamination of water, soil, and vegetables. BioMed Research International, vol. 2014, pp. 1-29. http://dx.doi.org/10.1155/2014/813206 PMid:25276818.
» http://dx.doi.org/10.1155/2014/813206 - WILSON, K., 2001. Preparation of genomic DNA from bacteria. Current Protocols in Molecular Biology, vol. 56, no. 15, pp. 2-4. https://doi.org/10.1002/0471142727.mb0204s56
» https://doi.org/10.1002/0471142727.mb0204s56 - WU, X., COBBINA, S.J., MAO, G., XU, H., ZHANG, Z. and YANG, L., 2016. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environmental Science and Pollution Research International, vol. 23, no. 9, pp. 8244-8259. http://dx.doi.org/10.1007/s11356-016-6333-x PMid:26965280.
» http://dx.doi.org/10.1007/s11356-016-6333-x - YASAR, A., KHAN, M., TABINDA, A.B., HAYYAT, M.U. and ZAHEER, A., 2013. Percentage uptake of heavy metals of different macrophytes in stagnant and flowing textile effluent. Journal of Animal and Plant Sciences, vol. 23, no. 1, pp. 1709-1713.
- ZEINER, M., REZIĆ, I. and STEFFAN, I., 2007. Analytical methods for the determination of heavy metals in the textile industry. Kemija u Industriji, vol. 56, no. 11, pp. 587-595.
Publication Dates
-
Publication in this collection
20 July 2020 -
Date of issue
Mar-May 2021
History
-
Received
10 Aug 2019 -
Accepted
14 Nov 2019