Abstract
Aflatoxin contamination of peanut, due to infection by Aspergillus flavus, is a major problem of rain-fed agriculture in India. In the present study, molecular characterisation of 187 Aspergillus flavus isolates, which were sampled from the peanut fields of Gujarat state in India, was performed using AFLP markers. On a pooled cluster analysis, the markers could successfully discriminate among the ‘A’, ‘B’ and ‘G’ group A. flavus isolates. PCoA analysis also showed equivalent results to the cluster analysis. Most of the isolates from one district could be clustered together, which indicated genetic similarity among the isolates. Further, a lot of genetic variability was observed within a district and within a group. The results of AMOVA test revealed that the variance within a population (84%) was more than that between two populations (16%). The isolates, when tested by indirect competitive ELISA, showed about 68.5% of them to be atoxigenic. Composite analysis between the aflatoxin production and AFLP data was found to be ineffective in separating the isolate types by aflatoxigenicity. Certain unique fragments, with respect to individual isolates, were also identified that may be used for development of SCAR marker to aid in rapid and precise identification of isolates.
aflatoxin; AMOVA; ELISA; genetic diversity; groundnut; PCA
Introduction
Peanut (Arachis hypogaea L.), also known as groundnut, is an important oilseed and ancillary food crop worldwide. In addition to the expulsion of oil, it is also used for production of peanut-butter and as a component of various food products. India possesses the largest peanut cultivation area in the world and is the second largest producer after China. The major Indian states, which collectively account for about 90% of the national area for peanut farming, include Gujarat, Andhra Pradesh, Tamil Nadu, Rajasthan, Karnataka and Maharashtra; Andhra Pradesh and Gujarat raking at first positions, in terms of cultivation area, and production, respectively. In Gujarat, about 80% of the peanut cultivation is concentrated in Junagadh, Rajkot, Porbandar, Amreli and Jamnagar districts of Saurashtra region (Anonymous, 2012Anonymous (2012) Annual Report, Directorate of Groundnut Research. Junagadh, Gujarat,, 136 pp.; Misra and Thirumalaisamy, 2012Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.).
Among the various parameters for quality assessment, aflatoxin contamination constitutes one of the major non-tariff trade barriers, especially, in the international peanut trade market (Misra and Thirumalaisamy, 2012Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.). Aflatoxins are secondary metabolites, which are produced by Aspergillus flavus group of fungi, and are known to be carcinogenic and mutagenic (Abbas et al., 2004Abbas HK, Shier WT, Horn BW et al. (2004) Cultural methods for aflatoxin detection. J Toxicol Toxin Rev 23:295–315.). Aflatoxin contamination of peanut, due to invasion by Aspergillus, is a major problem of the rain-fed agricultural cultivation conditions in India (Misra and Thirumalaisamy, 2012Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.). The fungi are wide spread in light sandy soils, which are most suitable for the peanut cultivation (Kumar et al., 2005Kumar V, Ghewande MP, Basu MS (2005) Safeguard Groundnut from Aflatoxin Contamination. NRCG, Junagadh, pp 1–13.). Although the aflatoxin contamination does not affect peanut production, but it causes serious health risks in humans and cattle (Horn et al., 1994Horn BW, Dorner JW, Greene RL et al. (1994) Effect of Aspergillus parasiticus soil inoculum on invasion of peanut seeds. Mycopathologia 125:179–191.).
Owing to the sensory properties, Indian peanuts are in great demand across the world. However, the export of peanut from India is hampered by aflatoxin contamination. The European Union (EU) has set a stringent maximum permissible limit (2 ppb) for aflatoxin in directly consumed peanuts (Wu et al., 2013Wu F, Stacy SL, Kensler TW (2013) Global Risk assessment of aflatoxins in maize and peanuts: Are regulatory standards adequately protective? TOXICOL SCI 135:251–259.). Due to aflatoxin contamination, recently, several consignments have been rejected at the destination ports in the EU (Misra and Thirumalaisamy, 2012Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.).
Peanut pods, when come in direct contact with the spores of A. flavus in soil, get frequently invaded before the harvest. The mode and extent of infection by the fungus depends on the population density of A. flavus in the soil, soil moisture and soil temperature during the pod development till maturity (Smith et al., 1995Smith JE, Solomons G, Lewis C et al. (1995) Role of mycotoxins in human and animal nutrition and health. Nat Toxins 3:187–192.). Once the kernels are contaminated, the elimination of aflatoxins is not possible by routine cooking or processing practices. Roasting, however, appreciably reduces the level of aflatoxin in the peanuts. Therefore, the best strategy to counteract this problem would be the prevention rather than decontamination (Misra and Thirumalaisamy, 2012Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.).
The aflatoxins are produced by the Aspergillus species, belonging to the section Flavi, such as, A. flavus, A. parasiticus and the others, like, A. nomius, A. minisclerotigenes, A. pseudocaelatus, etc. (Varga et al., 2011Varga J, Frisvad JC, Samson RA (2011) Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud Mycol 69:57–80.). Since, not all the isolates of Aspergillusare toxigenic (Desai et al., 1991Desai S, Ghewande MP, Nagaraj G et al. (1991) Screening for resistance to Aspergillus flavus and aflatoxin production in groundnut. Mycotoxin Res 7:79–84.), the characterisation of the isolates, for their toxigenicity in the major agro-ecological zones of peanut production system in India, is the need of the hour.
At present, DNA fingerprinting is relatively economical and allows discrimination of the fungal strains from the genus down to the clone level (Berbee and Taylor, 2001Berbee ML, Taylor JW (2001) Fungal Molecular Evolution: Gene Trees and Geologic Time. In: MC McLaughlin, EG McLaughlin, PA Lemke (eds) The Mycota VII Part B, Springer-Verlag, Berlin, pp 229–245.). Various types of molecular markers have been successfully employed to aid in detection of genetic variability in several Aspergillus species. Amplified Fragment Length Polymorphism (AFLP) is a technique which uses the benefits of both restriction digestion and PCR based selective amplification. AFLP has been widely used for molecular characterisation of Aspergillus spp. (Montiel et al., 2003Montiel D, Dickinson MJ, Lee HA et al. (2003) Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Res 107:1427–1434.; Lee et al., 2004Lee CZ, Liou GY, Yuan GF (2004) Comparison of Aspergillus flavus and Aspergillus oryzae by amplified fragment length polymorphism. Bot Bull Academia Sinica 45:61–68.). Till date, limited information, from India, is available on the prevalence and variability across the isolates of Aspergillus belonging to the section Flavi in respect to their toxigenicity (Desai et al., 1991Desai S, Ghewande MP, Nagaraj G et al. (1991) Screening for resistance to Aspergillus flavus and aflatoxin production in groundnut. Mycotoxin Res 7:79–84.; Rajarajan et al., 2013Rajarajan PN, Rajasekaran KM, Asha Devi NK (2013) Isolation and quantification of aflatoxin from Aspergillus flavus infected stored peanuts. Indian J Pharm Biol Res 1:76–80.). Hence, the present investigation was an attempt towards detailed molecular characterisation of A. flavus isolates, collected from different peanut cultivation fields in Gujarat. The ultimate aim of this study was to analyse the genetic association among the Aspergillus flavusisolates, with respect to their toxigenicity, in one of the major peanut producing set-up in India.
Materials and Methods
Fungal isolation and identification
A total of 187 fungal isolates, analysed in this study, were originally collected from the farmers’ fields, which were utilised for peanut farming, from 10 districts of Gujarat state (Table 1). Soil samples were collected from the groundnut fields and at each sampling, 5 randomly selected spots, at 0–10 cm of depth, from between the plants and individual samples were pooled for each plot. The interval between soil samples was 100–300 m at any single location, the pair-wise distance between populations was about 5–30 km, whereas, the pair-wise distance between the districts was approximately 50–430 km. The fungal isolation was done by the dilution plate method as previously described by Horn and Dorner (1998)Horn BW, Dorner JW (1998) Soil populations of Aspergillusspecies from section Flavi along a transect through peanut-growing regions of the United States. Mycologia 90:767–776. and the cultures were purified using the single spore isolation technique and maintained as single spore cultures on agar slants. All the isolates were cultured on Aspergillus flavus/parasiticus agar (AFPA; Sigma-Aldrich), which is a selective identification medium for the detection of A. flavus group strains, (Pitt et al., 1983Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54:109.) for 3 to 5 days at 25 °C in dark, to confirm identification at the section level by reverse colony colour. Further, the morphological and growth characteristics of all the isolates were analysed on solid medium, the Czapek’s Dox agar (CZ), and identification of the species was done on the basis of the colour of the colonies, i.e.yellow-green for A. flavus and dark green or nearly Ivy green for A. parasiticus.
Location and toxigenicity details of Aspergillus flavus isolates collected from soil samples under groundnut production system from Gujarat
Indirect competitive-enzyme linked immunosorbent assay (ELISA)
Indirect competitive ELISA was performed for the quantitative screening of the collected isolates. Aspergillus flavus strains were grown at 30 °C for 7 days on PDA plates (three replicates per isolate) as described by Waliyar et al. (2009)Waliyar F, Reddy SV, Lava-Kumar P (2009) Review of immunological methods for the quantification of aflatoxins in peanut and other foods. Peanut Sci 36:54–59.. Aflatoxin B1-bovine serum albumin (AFB1-BSA) conjugate was prepared in carbonate coating buffer (100 ng mL−1) and 150 μL was added to each well. The plates were then incubated at 37 °C for 1 h, after which the toxin was collected and stored. The wells were washed with PBST (Phosphate Buffered Saline supplemented with Tween 20) followed by incubation with BSA solution (0.2% BSA prepared in PBST) (200 μL per well) for 1 h at 37 °C. Antiserum diluted in BSA solution was added to the wells and incubated for 45 min at 37 °C. After appropriate blocking, the wells were washed with PBST.
Extract of healthy seed of peanut variety J-11 was taken as the negative control and for the positive control, the AFB1 standard was diluted (1:10) with peanut extract at concentrations ranging from 100 ng to 10 pg (100 μL per well). Then, 50 μL of the anti-serum (Sigma-Aldrich) was added to each dilution of aflatoxin standard (100 μL) and the peanut seed extract (100 μL). The plates having aflatoxin samples and antiserum were incubated at 37 °C for 1 h and subsequently washed with PBST.
Alkaline phosphatase (ALP) labelled goat anti-rabbit IgG (1:1000 dilution; volume 150 μL) was then added to each well and incubated at 37 °C for 1 h. The ELISA wells were washed with PBST and 150 μL of the substrate solution (p-nitrophenyl phosphate prepared in 10% diethanolamine buffer, pH 9.8) was added and incubated for 1 h at room temperature. The absorbance was measured at 405 nm in an automatic ELISA reader. A standard curve for AFB1 was prepared for estimation of aflatoxin content in the test samples. The detection limit for aflatoxin was 0.05 ppb.
Fungal DNA isolation
The isolates were cultured on potato dextrose agar (PDA) slants for isolation of genomic DNA. Conidia were harvested from 7-days old slant cultures, grown at 28 °C and inoculated into 50 mL of Yeast extract-Peptone-Dextrose broth followed by incubation at 25 °C for 48–72 h with shaking at150 rpm. After appropriate growth, the mycelial suspension was filtered through a Buchner funnel with sterile Whatman No. 1 filter paper. Mycelium was rinsed twice with sterile distilled water, transferred into a 50 mL centrifuge tube and froze at −80 °C.
Upon treatment with liquid nitrogen, the frozen mycelial mats were ground to fine powder using a mortar and pestle. Approximately, 20 mg of the homogenised mycelial powder was suspended in 600 μL of lysis buffer (100 mM Tris-HCl, pH 8.0; 100 mM NaCl; 20 mM EDTA and 2% SDS) and incubated for 10 min at 60 °C in a water bath. Subsequently, DNA was extracted from the samples by incubation with equal volumes of phenol/chloroform (1:1), followed by chloroform/isoamyl alcohol (24:1) treatment. DNA from the samples was precipitated with 0.7 volume of chilled ethanol and vacuum dried. Finally, the DNA pellets were re-suspended in TE Buffer (pH 8.0) and subjected to treatment with RNase A at 37 °C for 1.5 h, to remove RNA contamination. DNA concentration and purity was determined by measurement of absorbance at 260 nm and 280 nm using Nano Drop, while the integrity of DNA was examined by agarose gel (0.8%) electrophoresis.
AFLP reactions
AFLP analysis system for microorganisms (Invitrogen-Corporation, Carlsbad, CA) was used as previously described by Lee et al.(2004)Lee CZ, Liou GY, Yuan GF (2004) Comparison of Aspergillus flavus and Aspergillus oryzae by amplified fragment length polymorphism. Bot Bull Academia Sinica 45:61–68.. Approximately, 500 ng of genomic DNA, from each isolate, was subjected to restriction digestion with EcoRI and MseI restriction enzymes (Invitrogen-Corporation, Carlsbad, CA), and the restricted fragments were ligated to the double-stranded restriction site-specific ligation adaptors supplied with the kit. A pre-selective PCR (94 °C for 30 s, 20 cycles of 94 °C for 30 s, 56 °C for 60 s, 72 °C for 60 s; and 72 °C for 5 min; final hold at 4 °C) was carried out in a 25 μL (final volume) mixture. For the selective PCR, 5 μL of the 1:5 dilution of the first PCR product was amplified in a 25 μL (final volume) mixture using the selective primers.
Initially, 30 different primer combinations of EcoRI/MseI were used, of which five different primer combinations, viz. EcoRI-AA/Mse I-A, EcoRI-AC/Mse I-G, EcoRI-AC/Mse I-A, EcoRI-AC/Mse I-T and EcoRI-C/Mse I-CAG showed more polymorphism than the others and were used for the selective amplification. The PCR program, for selective AFLP amplification, included one cycle of 94 °C for 60 s and one cycle of 94 °C for 60 s, 65 °C for 60 s, and 72 °C for 90 s; this cycle was followed by nine cycles in which the annealing temperature ranged from 64 °C to 56 °C, and decreased by 1 °C for each cycle. Following that, 23 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s were performed, with the final extension at 72 °C for 5 min., and indefinite hold at 4 °C in a thermal cycler (Eppendorf). Thereafter, 5 μL of the reaction product was mixed with 3 μL of 6x loading dye (Fermentas). The amplification for each primer-combination was performed twice, independently, following the same procedure, in order to ensure the fidelity of the AFLP markers.
AFLP fragments were resolved in denaturing 6% polyacrylamide gel with 1X Tris-borate EDTA buffer (pH 8.0) in both the gels. The gels were run at 25 W and stained by silver staining with slight modifications, as described by Benbouza et al. (2006)Benbouza H, Jacquemin JM, Baudoin JP et al. (2006) Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol Agron Soc Environ 10:77–81., and scanned using the UMAX Mirage II gel scanner (Type H5K0). After digitisation of the gel pictures, the DNA bands were scored and analysed using the software Gel Compare II (Applied Maths, Kortrijk, Belgium).
Genetic distance and cluster analysis of AFLP data
Similarity matrix, using the AFLP polymorphism of A. flavus isolates, was measured by Jaccard similarity co-efficient, which was subjected to cluster analysis by Neighbour Joining method. FreeTree software (Pavlícek et al., 1999Pavlícek A, Hrdá S, Flegr J (1999) Free-Tree-freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha) 45:97–99.) was employed for construction of dendrogram, on the basis of distance data and for bootstrap analysis of the robustness of the trees. The colour separation in the dendrogram was done using the Interactive Tree of Life (Itol) software (Letunic and Bork, 2007Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128.). The allelic information was generated using the softwares, GenAIEx 6.501 and Gel Compare II. The AFLP data were subjected to a hierarchical analysis of molecular variance (AMOVA) (Excoffier et al., 1992Excoffier L, Smouse PE, Quattro JM (1992) Analyses of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491.), using the three hierarchical levels, i.e. individual, population and their regions. The GenAIEx software was used to calculate the principal co-ordinates analysis (PCoA) that plots the relationship between the distance matrix elements based on their first two principal co-ordinates (Peakall and Smouse, 2001Peakall R, Smouse PE (2001) GenAlEx V5: Genetic Analysis in Excel. Population genetic software for teaching and research. Australian National University, Canberra. http://www.anu.edu.au/BoZo/GenAlEx/.
http://www.anu.edu.au/BoZo/GenAlEx/...
).
Results and Discussion
Identification and toxigenicity of the isolates
In the present investigation, Aspergillus flavus populations were examined for their afla-toxigenicity. Amongst the isolates, belonging to Aspergillus section Flavi, 71% were characterised as A. flavus (n = 184) and the remaining 29% as A. parasiticus (n = 75). Our results are in concordance with the previous findings from studies on A. flavus population from the peanut cropping system in India (Patil 1985Patil SP, Shinde PA (1985) Mycotoxin contamination and associated mycoflora of groundnut. J Maharashtra Agric Univ 10:99–100., Reddy 2007Reddy BN, Raghavender CR (2007). Outbreaks of aflatoxicoses in India. Afr J Food Agril Nutr Develop 7:nd07046.). All the A. flavus isolates were selected as population and used for the AFLP analysis.
In this study, about 68.5% of the isolates were found to be atoxigenic, as tested by indirect competitive ELISA (Table 1), which is in conformity with the results of Chourasia and Sinha (1994)Chourasia HK, Sinha RK (1994) Potential of biological control of aflatoxin contamination in developing peanut by atoxigenic strains of Aspergillus flavus. J Food Sci Technol 31:362–366.. Bio-control by competitive exclusion has been regarded as the most promising means of controlling aflatoxin contamination of peanuts. It was observed that when competitive atoxigenic strains were applied to the soil, they produced large numbers of conidia than the toxigenic isolates (Alaniz Zanon et al., 2013Alaniz Zanon MS, Chiotta ML, Giaj-Merlera G et al. (2013) Evaluation of potential biocontrol agent for aflatoxin in Argentinean peanuts. Int J Food Microbiol 162:220–225.). Since, both occupy the same niches as the naturally occurring toxigenic populations, and aflatoxin contamination is subsequently reduced in the crops (Dorner, 2004Dorner JW (2004) Biological control of aflatoxin contamination of crops. J Toxicol Toxin Rev 23:425–450.). Such atoxigenic strains may be used for successful management of toxigenic Aspergilli in soil (Dorner and Lamb, 2006Dorner JW, Lamb MC (2006) Development and commercial use of aflaguard®, an aflatoxin biocontrol agent. Mycotoxin Res 21:33–38.).
Dendrogram analysis
In the present study, five selected AFLP primer-pair combinations produced a complex, but well-resolved fingerprint pattern (Figure 1). This analysis provided novel data on the molecular composition of A. flavuspopulations present in the peanut growing fields of Gujarat (India).
A composite dendrogram was generated based on all the five AFLP primer combinations using GenAIEx and Gel Compare II softwares, where all 187 isolates could be divided into 15 different clusters (I–XV; Figure 2). Moreover, the results of PCoA analysis were also comparable to the cluster analysis. Based on the morphological characterisation, the isolates were grouped into three distinct groups, i.e. group A, B and G. The dendrogram showed clear partitioning of ‘A’, ‘B’ and ‘G’ groups of the isolates into 14 (I–VIII and XII–XV), 02 (IX and X) and 01 (XI) clusters, respectively (Figure 2), which indicated that there were more scorable polymorphisms within the group ‘A’ isolates than either the ‘B’ or the ‘G’ groups of isolates. Our results are in agreement with Barros et al. (2007)Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909., who analysed the Aspergillus isolates by AFLP. Similar results have also been obtained through the other molecular methods (Tran Dinh, 1999Tran Dinh N, Pitt JI, Carter DA (1999) Molecular genotype analysis of natural toxigenic and nontoxigenic isolates of Aspergillus flavus and Aspergillus parasiticus. Mycol Res 103:1485–1490.; Wang et al., 2001Wang L, Yokoyama K, Takahasi H et al. (2001) Identification of species in Aspergillus section Flavi based on sequencing of mitochondrial cytochrome b gene. Int J Food Microbiol 71:75–86.).
Clustering pattern of 187 A. flavus isolates based on AFLP analysis. Where, group ‘A’, ‘B’ and ‘G’ isolates were clustered in 12 (I–VIII and XII–XV); 02 (IX–X) and 01 (XI) clusters respectively
From the cluster pattern based on the composite AFLP analysis, it has been inferred that most of the isolates of the same district could be clustered together. A few isolates of the other districts, like, Junagadh were also found to be clustered with the isolates of the other districts. This might be due to the dissemination of the contaminated seed material, being cultivated in one district, to another or it could be because of a similar genetic make-up of those isolates. Such result agrees with a number of previous findings in Flavi section of Aspergillus (Montiel et al., 2003Montiel D, Dickinson MJ, Lee HA et al. (2003) Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Res 107:1427–1434.; Barros et al., 2007Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909.; Baird et al., 2006Baird RE, Trigiano RN, Windham G et al. (2006) Comparison of aflatoxigenic and nonaflatoxigenic isolates of Aspergillus flavususing DNA amplification fingerprinting techniques. Mycopathologia 161:93–99.). Thus, by using these five AFLP primer combinations, the A. flavus isolates could be grouped according to their morphological groups (e.g. Group ‘A’, ‘B’ and ‘G’) and, to some extent, to their geographical location too.
Details of genetic diversity of different A. flavuspopulations
While analysing the genetic diversity details of each population of A. flavus, it was observed that the maximum number of bands were amplified, and percentage polymorphic bands were recorded for Junagadh district (360 and 99.17%, respectively), followed by Amreli district (325 and 89.26%, respectively). Likewise, the number of different alleles and the number of effective alleles were also the highest for the isolates that were collected from the Junagadh district. This clearly indicated that the maximum diversity was recorded for Junagadh and minimum for Anand district (Table 2). Certain differences could be attributed to the size of fungal population that was studied from any location, i.e.the larger is the population, more pronounced are its genetic diversity details. Genetic diversity details of all the populations studied have been presented in the Table 2.
Genetic diversity details about each population of A. flavuscollected from different parts of groundnut growing fields of Gujarat
AMOVA revealed that the variance within the population was much more (84%) than that between the populations (16%). A higher genetic diversity within the A. flavus populations indicates that there could be many discreet populations that possess unique genotypes at any location. However, on the population basis, at any two locations, variations are not so high. Thus, high level of genetic diversity observed within the populations of A. flavus could be attributed to the evolutionary factors, such as gene-flow, random genetic drift and the anthropogenic activities, such as a specific peanut cropping pattern followed in Gujarat, which needs further investigation. The results of various previous studies on A. flavus (Montiel et al., 2003Montiel D, Dickinson MJ, Lee HA et al. (2003) Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Res 107:1427–1434.; Barros et al., 2007Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909.; Baird et al., 2006Baird RE, Trigiano RN, Windham G et al. (2006) Comparison of aflatoxigenic and nonaflatoxigenic isolates of Aspergillus flavususing DNA amplification fingerprinting techniques. Mycopathologia 161:93–99.) are in tune with our investigation findings.
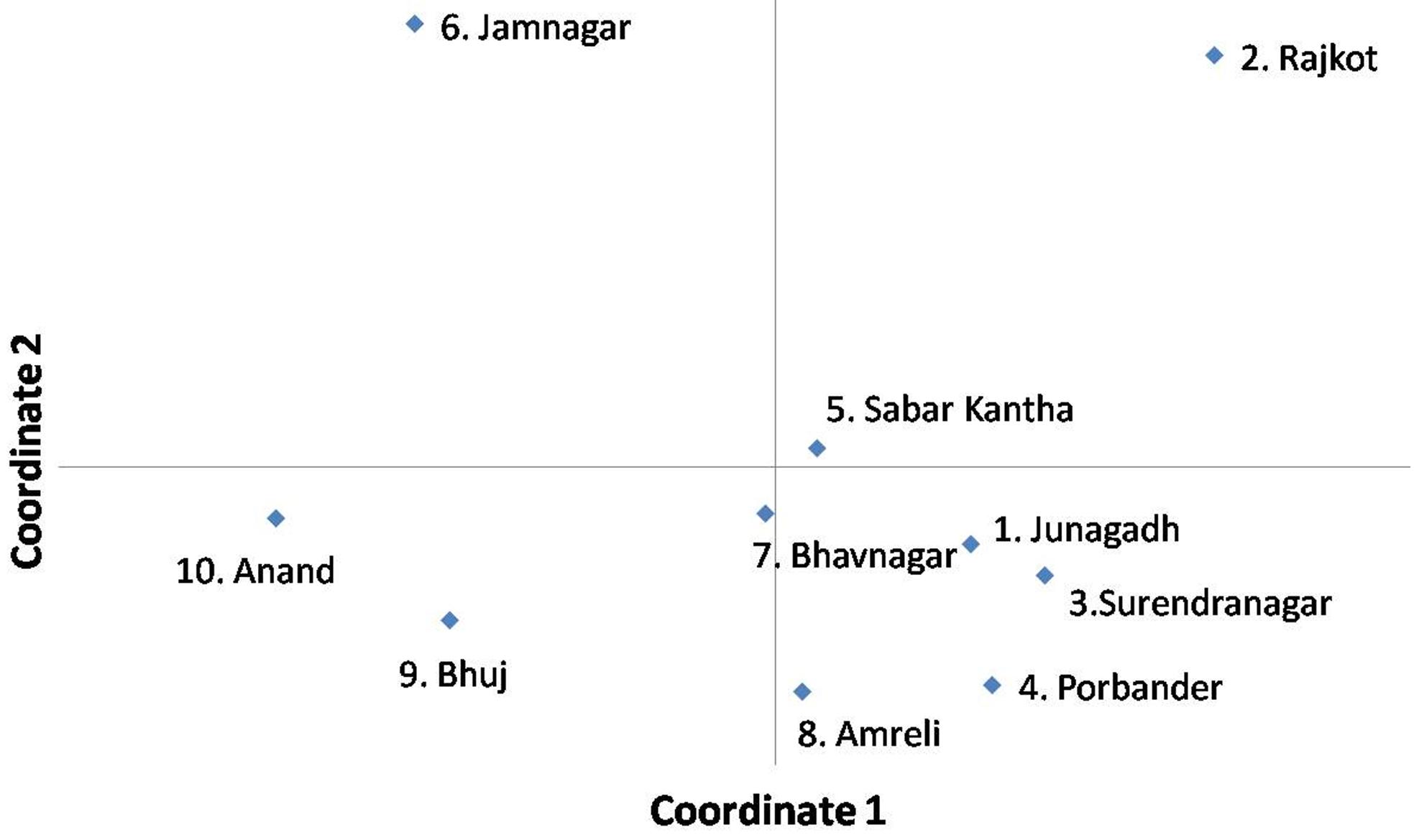
Analysis of the principal co-ordinates of A. flavus populations, which were collected from the fields of 10 peanut growing districts of Gujarat, revealed that the isolates collected from the districts of Junagadh, Sabar Kantha, Surendranagar, Bhavnagar, Porbander and Amreli are closer to each other. However, the isolates collected from Jamnagar, Rajkot, Anand and Bhuj district are quite diverse from each other (Figure 3).
Principal coordinates analysis of A. flavus populations, collected from the fields of ten peanut growing districts of Gujarat state
Identification of different groups of isolates
The AFLP primer combinations used in the present study showed certain specific fragments, which may be specifically used for the identification of isolates from each other. For certain isolates, 14 unique bands were identified (Table 3). The 200 bp fragment, amplified by E-AA/M-A primer combination, was absent in each of the ‘A’ group isolates, whereas, it was present in all of the ‘B’ and the ‘G’ group isolates. A fragment of 400 bp, amplified by E-AC/M-A primer combination, was present in all the ‘G’ group isolates, but absent in all the ‘B’ and the ‘A’ group isolates. The same primer combination could amplify a fragment of 525 bp that was present only in the ‘B’ group isolates and absent in all the ‘A’ and the ‘G’ group isolates. Some unique fragments, which were either present or absent in isolates, may also serve as specific markers for identification of respective isolates or group of isolates.
Amplicons that can be used for SCAR development against specific isolates and/or group of isolates
Some specific isolates, i.e. NRCG 02037, 02021 and 05024, may be identified by using the specific primer combinations. Apart from the individual isolates, a set of isolates and a group of isolates have also produced specific fingerprint and can be identified by using the primer combinations used in our study. Group ‘B’, ‘G’ and ‘A’ isolates produced different fingerprints and thus, could be differentiated from each other. Since, the PCR diagnostics could be of great value in ecological and epidemiological studies, where vast numbers of isolates have to be screened in a short duration (Schmidt et al., 2004Schmidt H, Taniwaki MH, Vogel RF et al. (2004) Utilization of AFLP markers for PCR-based identification of Aspergillus carbonariusand indication of its presence in green coffee samples. J Appl Microbiol 97:899–909.), therefore these diagnostic/specific fragments could be useful in establishment of a PCR-based diagnostic assay (Barros et al., 2007Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909.), by development of sequence characterised amplified regions (SCARs). In the current investigation, certain specific amplicons were identified which may be used for the development of SCARs for identification of specific isolates (Table 3).
Association of AFLP markers with toxigenicity
The molecular mechanisms leading to the loss of aflatoxin production in atoxigenic A. flavus have been investigated intensively by various researchers across the world (Jiang et al., 2009Jiang J, Yan L, Ma Z (2009) Molecular characterization of an atoxigenic Aspergillus flavus strain AF051. Appl Microbiol Biotechnol DOI 10.1007/s00253-009-1921-z.
https://doi.org/10.1007/s00253-009-1921-...
; Criseo et al., 2008Criseo G, Racco C, Romeo O (2008) High genetic variability in nonaflatoxigenic A. flavus strains by using quadruplex PCR-based assay. Int J Food Microbiol 125:341–343.). In the present investigation, AFLP analysis was found to be ineffective for differentiation of isolate types on the basis of aflatoxigenicity, as both toxigenic and atoxigenic forms were inter-mixed within the groups with no clear demarcation. Our result is in concurrence with the previous studies on Aspergillus section Flavi, in view of RAPDs (Tran Dinh, 1999Tran Dinh N, Pitt JI, Carter DA (1999) Molecular genotype analysis of natural toxigenic and nontoxigenic isolates of Aspergillus flavus and Aspergillus parasiticus. Mycol Res 103:1485–1490.), quadruplex PCR (Wang et al., 2001Wang L, Yokoyama K, Takahasi H et al. (2001) Identification of species in Aspergillus section Flavi based on sequencing of mitochondrial cytochrome b gene. Int J Food Microbiol 71:75–86.), AFLP (Montiel et al., 2003Montiel D, Dickinson MJ, Lee HA et al. (2003) Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Res 107:1427–1434.), and DNA amplification fingerprinting (Baird et al., 2006Baird RE, Trigiano RN, Windham G et al. (2006) Comparison of aflatoxigenic and nonaflatoxigenic isolates of Aspergillus flavususing DNA amplification fingerprinting techniques. Mycopathologia 161:93–99.). Moreover, the earlier studies have also shown that with AFLP, no genotypic difference could be established between the toxin producers and the non-producers (Barros et al., 2007Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909.; Schmidt et al., 2004Schmidt H, Taniwaki MH, Vogel RF et al. (2004) Utilization of AFLP markers for PCR-based identification of Aspergillus carbonariusand indication of its presence in green coffee samples. J Appl Microbiol 97:899–909.; Perrone et al., 2006Perrone G, Mule G, Susca A et al. (2006) Ochratoxin A production and amplified fragment length polymorphism analysis of Aspergillus carbonarius, Aspergillus tubigiensis and Aspergillus nigerstrains isolated from grapes in Italy. Appl Environ Microbiol 72:680–685.).
As toxin production is a very complex trait and is unlikely to be acquired independently, Tran et al. (1999)Tran Dinh N, Pitt JI, Carter DA (1999) Molecular genotype analysis of natural toxigenic and nontoxigenic isolates of Aspergillus flavus and Aspergillus parasiticus. Mycol Res 103:1485–1490. suggested that in the absence of sexual recombination, non-toxigenicity has been lost multiple times by different isolates. Geiser et al.(1998)Geiser DM, Pitt JI, Taylor JW (1998) Crytic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. P Natl Acad Sci USA 95:388–393. proved the recombination in A. flavus, which means, non-toxigenicity may have passed laterally between isolates of different genetic backgrounds.
It has been reported that the analysis of deletions within the aflatoxin biosynthetic gene cluster, could be a more effective marker for differentiation of toxigenic and atoxigenic isolates (Jiang et al., 2009Jiang J, Yan L, Ma Z (2009) Molecular characterization of an atoxigenic Aspergillus flavus strain AF051. Appl Microbiol Biotechnol DOI 10.1007/s00253-009-1921-z.
https://doi.org/10.1007/s00253-009-1921-...
; Chang et al., 2005Chang PK, Horn BW, Dorner JW (2005) Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol 42:914–923.). However, the loss of aflatoxin production may be not result only due to deletions in the gene cluster (Wang et al., 2001Wang L, Yokoyama K, Takahasi H et al. (2001) Identification of species in Aspergillus section Flavi based on sequencing of mitochondrial cytochrome b gene. Int J Food Microbiol 71:75–86.; Criseo et al., 2008Criseo G, Racco C, Romeo O (2008) High genetic variability in nonaflatoxigenic A. flavus strains by using quadruplex PCR-based assay. Int J Food Microbiol 125:341–343.). Criseo et al. (2008)Criseo G, Racco C, Romeo O (2008) High genetic variability in nonaflatoxigenic A. flavus strains by using quadruplex PCR-based assay. Int J Food Microbiol 125:341–343. reported that 36.5% of atoxigenic strains have the complete aflatoxin gene cluster; however, the exact mechanism of loss of aflatoxin production is still unknown. In the atoxigenic strain AF36, a defect causing a premature stop codon in the coding sequence of the aflatoxin biosynthesis gene pksA was reported (Ehrlich and Cotty, 2004Ehrlich KC, Cotty PJ (2004) An isolate of Aspergillus flavus used to reduce aflatoxin contamination in cotton seed has a defective polyketide synthase gene. Appl Microbiol Biotechnol 65:473–478.). This suggests that there is a need to further characterise the non-aflatoxigenic strains, which have been identified in the present investigation, to find the exact cause of their atoxigenicity.
Based on our findings, we may conclude that the AFLP technique can provide the required genetic information about the A. flavus isolates from the peanut cropping systems in India. Moreover, it can also be used as a powerful molecular tool to study the genetic diversity in A. flavus. The information generated, from this study, could be used for the prevention of aflatoxin contamination of peanut crop, at field level, by increasing the relative concentration of the atoxigenic strains, in one of the major peanut cultivation area in India, which will ultimately help in improving the crop’s export. Further, the molecular characterisation of atoxigenic strains that were identified in the study would be useful to unveil the basis of their atoxigenicity.
Acknowledgments
Authors are thankful to the ICAR, New Delhi for funding of the project under the “Network project on Prevention and management of mycotoxin contamination in commercially important agricultural commodities”. We thank “Sci-Edit Publications (Language Editing Services) (www.sci-edit.net)” for editing the manuscript.
References
- Abbas HK, Shier WT, Horn BW et al. (2004) Cultural methods for aflatoxin detection. J Toxicol Toxin Rev 23:295–315.
- Alaniz Zanon MS, Chiotta ML, Giaj-Merlera G et al. (2013) Evaluation of potential biocontrol agent for aflatoxin in Argentinean peanuts. Int J Food Microbiol 162:220–225.
- Anonymous (2012) Annual Report, Directorate of Groundnut Research. Junagadh, Gujarat,, 136 pp.
- Baird RE, Trigiano RN, Windham G et al. (2006) Comparison of aflatoxigenic and nonaflatoxigenic isolates of Aspergillus flavususing DNA amplification fingerprinting techniques. Mycopathologia 161:93–99.
- Barros GG, Chiotta ML, Reynoso MM et al. (2007) Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J Appl Microbiol 103:900–909.
- Benbouza H, Jacquemin JM, Baudoin JP et al. (2006) Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol Agron Soc Environ 10:77–81.
- Berbee ML, Taylor JW (2001) Fungal Molecular Evolution: Gene Trees and Geologic Time. In: MC McLaughlin, EG McLaughlin, PA Lemke (eds) The Mycota VII Part B, Springer-Verlag, Berlin, pp 229–245.
- Chang PK, Horn BW, Dorner JW (2005) Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol 42:914–923.
- Chourasia HK, Sinha RK (1994) Potential of biological control of aflatoxin contamination in developing peanut by atoxigenic strains of Aspergillus flavus J Food Sci Technol 31:362–366.
- Criseo G, Racco C, Romeo O (2008) High genetic variability in nonaflatoxigenic A. flavus strains by using quadruplex PCR-based assay. Int J Food Microbiol 125:341–343.
- Desai S, Ghewande MP, Nagaraj G et al. (1991) Screening for resistance to Aspergillus flavus and aflatoxin production in groundnut. Mycotoxin Res 7:79–84.
- Dorner JW (2004) Biological control of aflatoxin contamination of crops. J Toxicol Toxin Rev 23:425–450.
- Dorner JW, Lamb MC (2006) Development and commercial use of aflaguard®, an aflatoxin biocontrol agent. Mycotoxin Res 21:33–38.
- Ehrlich KC, Cotty PJ (2004) An isolate of Aspergillus flavus used to reduce aflatoxin contamination in cotton seed has a defective polyketide synthase gene. Appl Microbiol Biotechnol 65:473–478.
- Excoffier L, Smouse PE, Quattro JM (1992) Analyses of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491.
- Geiser DM, Pitt JI, Taylor JW (1998) Crytic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus P Natl Acad Sci USA 95:388–393.
- Horn BW, Dorner JW (1998) Soil populations of Aspergillusspecies from section Flavi along a transect through peanut-growing regions of the United States. Mycologia 90:767–776.
- Horn BW, Dorner JW, Greene RL et al. (1994) Effect of Aspergillus parasiticus soil inoculum on invasion of peanut seeds. Mycopathologia 125:179–191.
- Jiang J, Yan L, Ma Z (2009) Molecular characterization of an atoxigenic Aspergillus flavus strain AF051. Appl Microbiol Biotechnol DOI 10.1007/s00253-009-1921-z.
» https://doi.org/10.1007/s00253-009-1921-z - Kumar V, Ghewande MP, Basu MS (2005) Safeguard Groundnut from Aflatoxin Contamination. NRCG, Junagadh, pp 1–13.
- Lee CZ, Liou GY, Yuan GF (2004) Comparison of Aspergillus flavus and Aspergillus oryzae by amplified fragment length polymorphism. Bot Bull Academia Sinica 45:61–68.
- Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128.
- Misra JB, Thirumalaisamy PP (2012) Bulletin on Management of Aflatoxin. Directorate of Groundnut Research, Junagadh, 52 pp.
- Montiel D, Dickinson MJ, Lee HA et al. (2003) Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Res 107:1427–1434.
- Patil SP, Shinde PA (1985) Mycotoxin contamination and associated mycoflora of groundnut. J Maharashtra Agric Univ 10:99–100.
- Pavlícek A, Hrdá S, Flegr J (1999) Free-Tree-freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha) 45:97–99.
- Peakall R, Smouse PE (2001) GenAlEx V5: Genetic Analysis in Excel. Population genetic software for teaching and research. Australian National University, Canberra. http://www.anu.edu.au/BoZo/GenAlEx/
» http://www.anu.edu.au/BoZo/GenAlEx/ - Perrone G, Mule G, Susca A et al. (2006) Ochratoxin A production and amplified fragment length polymorphism analysis of Aspergillus carbonarius, Aspergillus tubigiensis and Aspergillus nigerstrains isolated from grapes in Italy. Appl Environ Microbiol 72:680–685.
- Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54:109.
- Rajarajan PN, Rajasekaran KM, Asha Devi NK (2013) Isolation and quantification of aflatoxin from Aspergillus flavus infected stored peanuts. Indian J Pharm Biol Res 1:76–80.
- Reddy BN, Raghavender CR (2007). Outbreaks of aflatoxicoses in India. Afr J Food Agril Nutr Develop 7:nd07046.
- Schmidt H, Taniwaki MH, Vogel RF et al. (2004) Utilization of AFLP markers for PCR-based identification of Aspergillus carbonariusand indication of its presence in green coffee samples. J Appl Microbiol 97:899–909.
- Smith JE, Solomons G, Lewis C et al. (1995) Role of mycotoxins in human and animal nutrition and health. Nat Toxins 3:187–192.
- Tran Dinh N, Pitt JI, Carter DA (1999) Molecular genotype analysis of natural toxigenic and nontoxigenic isolates of Aspergillus flavus and Aspergillus parasiticus Mycol Res 103:1485–1490.
- Varga J, Frisvad JC, Samson RA (2011) Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud Mycol 69:57–80.
- Waliyar F, Reddy SV, Lava-Kumar P (2009) Review of immunological methods for the quantification of aflatoxins in peanut and other foods. Peanut Sci 36:54–59.
- Wang L, Yokoyama K, Takahasi H et al. (2001) Identification of species in Aspergillus section Flavi based on sequencing of mitochondrial cytochrome b gene. Int J Food Microbiol 71:75–86.
- Wu F, Stacy SL, Kensler TW (2013) Global Risk assessment of aflatoxins in maize and peanuts: Are regulatory standards adequately protective? TOXICOL SCI 135:251–259.
Publication Dates
-
Publication in this collection
21 July 2015 -
Date of issue
Jul-Sep 2015
History
-
Received
19 Oct 2013 -
Accepted
12 Nov 2014