Abstract
Emergence and distribution of multi-drug resistant (MDR) bacteria in environments pose a risk to human and animal health. A total of 82 isolates of Escherichia spp. were recovered from cloacal swabs of migrating and non-migrating wild birds. All bacterial isolates were identified and characterized morphologically and biochemically. 72% and 50% of isolates recovered from non-migrating and migrating birds, respectively, showed positive congo red dye binding (a virulence factor). Also, hemolysin production (a virulence factor) was showed in 8% of isolates recovered from non-migrating birds and 75% of isolates recovered from migrating birds. All isolates recovered from non-migrating birds were found resistant to Oxacillin while all isolates recovered from migrating birds demonstrated resistance to Oxacillin, Chloramphenicol, Oxytetracycline and Lincomycin. Some bacterial isolates recovered from non-migrating birds and migrating birds exhibited MDR phenotype. The MDR isolates were further characterized by API 20E and 16S rRNA as E. coli and E. vulneris. MDR Escherichia isolates contain ~1-5 plasmids of high-molecular weights. Accordingly, wild birds could create a potential threat to human and animal health by transmitting MDR bacteria to water streams and other environmental sources through their faecal residues, and to remote regions by migration.
E. coli; E. vulneris; multi-drug resistance (MDR); migrating and non-migrating birds; congo red binding; hemolysin; API 20E; 16S rRNA; plasmid profile
RESEARCH PAPER
Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris
Mohammed Y. ShobrakI; Aly E. Abo-AmerII, III
IDivision of Zoology, Faculty of Science, University of Taif, Taif, Saudi Arabia
IIDivision of Microbiology, Department of Biology, Faculty of Science, University of Taif, Taif, Saudi Arabia
IIIDivision of Microbiology, Department of Botany, Faculty of Science, Sohag University, Sohag, Egypt
Correspondence Correspondence: A.E. Abo-Amer Division of Microbiology Department of Biology Faculty of Science Taif University 888 Taif, Saudi Arabia E-mail: abo-amer@lycos.com
ABSTRACT
Emergence and distribution of multi-drug resistant (MDR) bacteria in environments pose a risk to human and animal health. A total of 82 isolates of Escherichia spp. were recovered from cloacal swabs of migrating and non-migrating wild birds. All bacterial isolates were identified and characterized morphologically and biochemically. 72% and 50% of isolates recovered from non-migrating and migrating birds, respectively, showed positive congo red dye binding (a virulence factor). Also, hemolysin production (a virulence factor) was showed in 8% of isolates recovered from non-migrating birds and 75% of isolates recovered from migrating birds. All isolates recovered from non-migrating birds were found resistant to Oxacillin while all isolates recovered from migrating birds demonstrated resistance to Oxacillin, Chloramphenicol, Oxytetracycline and Lincomycin. Some bacterial isolates recovered from non-migrating birds and migrating birds exhibited MDR phenotype. The MDR isolates were further characterized by API 20E and 16S rRNA as E. coli and E. vulneris. MDR Escherichia isolates contain ~1-5 plasmids of high-molecular weights. Accordingly, wild birds could create a potential threat to human and animal health by transmitting MDR bacteria to water streams and other environmental sources through their faecal residues, and to remote regions by migration.
Key words: E. coli, E. vulneris, multi-drug resistance (MDR), migrating and non-migrating birds, congo red binding, hemolysin, API 20E, 16S rRNA, plasmid profile.
Introduction
During the time of World War II, antimicrobial agents were first widely utilized to treat bacterial infections. Since that time, antimicrobial agent use has gradually increased. In addition to huge use in the treatment of human illnesses, antibiotics are commonly used in veterinary medicine and usually added to the nourish of food producing animals. Random use of antimicrobial agents may cause existing populations of microorganisms that are antimicrobial resistant or multi-drug resistant (MDR). These microorganisms may be shed in faeces with following contamination of soil, food, and aquatic environments. While antimicrobial agents have confirmed a successful way against bacterial contamination and infection, their widespread use has produced a reservoir of antimicrobial agents and MDR microorganisms. The occurrence and persistence of antimicrobial resistant bacteria in such environments as sediment (Timoney et al., 1978), soil (Trevors, 1987), surface water (Bell et al., 1980; Wnorowski, 1993), municipal drinking water (Armstrong et al., 1981; Calomiris et al., 1984; Moffie and Mouton, 1988) and sewage (Altherr and Kasweck, 1982; Middleton and Ambrose, 2005; Murray et al., 1984; Walter and Vennes, 1985), is a growing public health concern.
The appearance of MDR bacteria of animals, birds and human (Grobbel et al., 2007) is accompanied by co-contamination of the environment leading to a great health alarm (Martinez, 2009). In addition to the present detection of MDR bacteria in high human density sites (Cole et al., 2005), their incidence in more distant regions like high mountains or the arctic is more concern (Caprioli et al., 1991; Sjolund et al., 2008). The presence of bacteria of potential zoonotic importance among migrating and non-migrating wild birds has public health significance. Different of pathogenic bacterial species have been isolated from wild birds. Migrating and non-migrating wild birds can also act as reservoirs of coliform bacteria, such as E. coli, carrying antimicrobial-resistance genes. Water contact and acquirement by food seem to be the main factors of transmission of resistant bacteria from human or veterinary origin to wild animals (Cole et al., 2005; Kozak et al., 2009). Migrating and non-migrating wild birds or general wild animals could therefore serve as reservoirs of resistant bacteria and genetic factors of antimicrobial resistance (Dolejska et al., 2009).
Antimicrobial resistance has been known as an emerging worldwide problem in both human and veterinary medicine, and antimicrobial use is considered the most important factor for the emergence, selection, and distribution of antimicrobial-resistant bacteria (Winokur et al., 2001). Enteropathogenic strains, such as the vero cytotoxin-producing E. coli O157:H7 strain, are the agent of colibacillosis and have been isolated from healthy or diseased wild birds, including migrants such as Ardea cinerea, B. canadensis, Cygnus columbianus, U. aalge, and Columba palumbus (Hubalek, 1994). These migrating birds can become carriers of E. coli strains resistant to antimicrobial agents and can be responsible for the spread of R plasmids over the world (Kanai et al., 1981). Migratory waterfowl may serve as reservoirs of antimicrobial resistance of thermotolerant faecal indicator organisms (Middleton and Ambrose, 2005). E. coli isolates originating from Arctic birds carry antimicrobial resistance determinants.
The problem of antimicrobial resistance worldwide is one of the foremost issues that we face in the coming decades. We strongly believe that there is an urgent requirement for research on how to comprehensively address the problems of antimicrobial resistance. The problem of resistance as a public health threat has increased significantly over the last decade and local solutions are needed. The aim of this study was to obtain detailed understanding of possible migrating and non-migrating wild birds carry and spread of drug-resistant Escherichia spp. in the environment which could form a potential hazard to human and animal health by transmission of antimicrobial-resistant strains to waterways and environmental sources through faecal deposits.
Therefore, this work was approached to isolate, characterize and verify the prevalence of antimicrobial resistant Escherichia spp. in migrating and non-migrating wild birds with an attention on the detection of MDR bacteria.
Materials and Methods
Sampling sites and birds capture
Fifteen different types of migrating and non-migrating wild birds (Three of each species) were captured (Table 1), using misnets and clap-nets, from different areas (Jabra; Meisan, Samnan Wa Samneen, National Wildlife Research Center (near Besel village, Wadi Jaleel and Wadi Al Arj) at Taif province, Saudi Arabia. Birds were captured within a vegetated area covering 500 m radius. Birds were characterized and identified according to Porter and Aspinall (Porter and Aspinall, 2010). Capturing the wild birds was approved by the Saudi Wildlife Authority, Taif province.
Sampling from cloacal swabs
Sterile cotton swabs wetted with sterile normal saline water were inserted in the cloacae of the migrating and non-migrating wild birds, and placed in sterile vials. After collection of cloacal swabs the birds were then freed. The samples were transported immediately to the laboratory in an ice box.
Isolation and identification of Escherichia spp.
Standard methods were used for the enrichment, isolation, identification, and biochemical confirmation of Escherichia spp. isolates (Clesceri et al., 1998). All samples were processed within 4 h. Swab samples were enriched in buffered peptone water at 37 °C for 24 h. Subsequently, the cultures were streaked on MacConkey agar and incubated at 37 °C overnight. The plates were investigated for lactose fermenting bacteria (red colonies) that precipitated bile and had a dark red center. Different single colonies were then collected and purified.
All isolates were morphologically characterized by Gram stain. For biochemical characterization, isolates were inoculated in tryptic soy broth (TSB), and incubated for approximately 4 h until the cultures were turbid. Identification was done according to Buchanan and Gibbons (1974) following a series of biochemical tests included oxidase, methyl red, Voges-Proskauer reactions, indole, citrate, catalase, urea hydrolysis, gelatin hydrolysis, lactose fermentation, nitrate reduction, casein hydrolysis and sugar fermentation. Moreover, identification of Escherichia spp. isolates was further confirmed by analytical profile index API 20E strips (Bio Merieux).
Congo red binding
To estimate the Congo red dye binding (Styles and Flammer, 1991), Escherichia spp. isolates were grown at 37 °C for 24 h on tryptic soy agar supplemented with 0.02% Congo red (Sigma) and 0.15% bile salt (Difco). Positive colonies were red, while negative colonies were appeared pale. Based on the intensity of red color, the binding was recorded as +, ++, and +++.
Hemolysis
To investigate hemolysis (Hacker et al., 1983), overnight cultures of Escherichia spp. isolates were streaked on sheep blood agar and incubated at 37 °C for 24 h. The appearance of a zone of erythrocyte lysis around or under bacterial colonies indicated hemolysis.
Antimicrobial susceptibility test
Only the bacterial isolates that confirmed to be Escherichia spp. based on the results of the biochemical tests were selected for antimicrobial agent sensitivity testing. The antimicrobial sensitivity phenotypes of Escherichia spp. isolates were determined using a Kirby-Bauer disk diffusion assay according to the standards and interpretive criteria described by CLSI (CLSI, 2012).
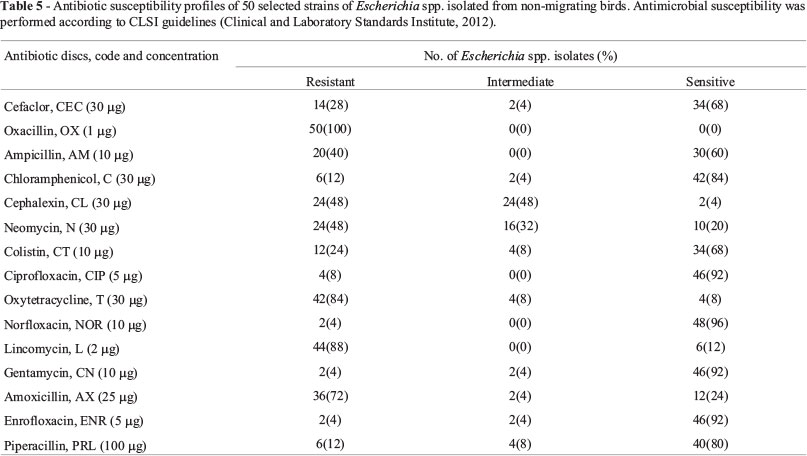
A total of 15 antimicrobial discs with Cefaclor (30 µg), Oxacillin (1 µg), Ampicillin (10 µg), Chloramphenicol (30 µg), Cephalexin (30 µg), Neomycin (30 µg), Colistin (10 µg), Ciprofloxacin (5 µg), Oxytetracycline (30 µg), Norfloxacin (10 µg), Lincomycin (2 µg), Gentamycin (10 µg), Amoxicillin (25 µg), Enrofloxacin (5 µg) and Piperacillin (100 µg) were used. This antimicrobial panel was chosen to include antibiotics with potential efficiency against Escherichia spp. isolates. These antimicrobial agents were chosen on the basis of their importance in treating human or animal E. coli infections and their use as feed additives to promote growth in animals and on the basis of their ability to provide diversity for representation of different antimicrobial classes (Krumperman, 1983).
Plates of Mueller-Hinton medium were swabbed with TSB broth inoculated with Escherichia spp. isolates and incubated to a turbidity of 0.5 McFarland standard. The commercially prepared antimicrobial disks (4x50, BIO-RAD) were placed on the inoculated plates. The plates were incubated at 35 °C for 20 h. The diameters (millimeters) of the clear zones of growth inhibition around the antimicrobial agent disks, including the 6-mm disk diameter, were measured by using precision calipers. The zone diameter for individual antimicrobial agents was then translated into sensitive, intermediate and resistant categories according to the interpretation table of the CLSI (CLSI, 2012). Multi-drug resistance (MDR) refers to resistance to 3 or more antimicrobials.
Molecular identification of the most MDR bacteria
Preparation of genomic DNA
Bacterial colonies were picked up with a sterilized toothpick, and suspended in 0.5 mL of sterilizes saline in a 1.5 mL centrifuge tube. Centrifugation was performed at 10,000 rpm for 10 min. After removal of supernatant, the pellet was suspended in 0.5 mL of InstaGene Matrix (Bio-Rad, USA) and incubated 56 °C for 30 min and then heated 100 °C for 10 min. After heating, supernatant was used for PCR.
PCR of 16S rRNA gene
One microliter of template DNA was added in 20 L of PCR reaction solution. 27F (AgA gTT TgA TCM TGG CTC Ag) and 1492R (TAC ggY TAC CTT gTT ACg ACT T) primers were used. 35 amplification cycles were performed at 94 °C for 45 s, 55 °C for 60 s, and 72 °C for 60 s. DNA fragments were amplified ~ 1,400 bp.
Unincorporated PCR primers and dNTPs were removed from PCR products by using Montage PCR Clean up kit (Millipore).
Sequencing
The purified PCR products of approximately 1,400 bp were sequenced by using two primers which were 518F (CCA gCA gCC gCg gTA ATA Cg) and 800R (TAC CAg ggT ATC TAA TCC). Sequencing was performed by using Big Dye terminator cycle sequencing kit (Applied BioSystems, USA). Sequencing products were resolved on an Applied Biosystems model 3730XL automated DNA sequencing system (Applied BioSystems, USA).
Selected sequences of other microorganisms with greatest similarity to the 16S rRNA sequences of bacterial isolates were extracted from the nucleotide sequence databases and aligned using CLUSTAL W (1.81) Multiple Sequence Alignment generating phylogenetic tree.
The 16S rRNA gene sequences of the bacterial isolates reported in this paper were deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession numbers: AB758349 (E. coli WB3-1), AB758350 (E. coli WB3-2), AB758351 (E. coli MB17-1), AB758352 (E. coli MB17-2), AB758353 (E. vulneris WB7-2), AB758354 (E. vulneris MB14-1), AB758355 (E. vulneris MB14-2), AB758356 (E. vulneris MB21-1) and AB758357 (E. vulneris MB21-2).
Plasmid analysis
Plasmid extraction of MDR isolates was carried out by alkaline lysis technique (Anderson and McKa, 1983). The isolated plasmid was then separated using a horizontal 1% agarose gel electrophoresis technique. Shigella flexneri containing plasmids of known molecular weights was used as a standard. The log of the molecular weights (in base pairs) vs. the distances (cm) traveled for the standard plasmids was plotted and then the sizes of unknown plasmids were estimated.
Curing of plasmid
Plasmid curing was achieved for MDR isolates by physical method by incubating bacterial cells at 45 °C as reported by Fortina and Silva (1996). The bacterial isolates were incubated in TSB at 45 °C overnight for plasmid curing. Other flasks were incubated at 37 °C as a control. The sodium dodecyl sulphate (1% SDS) treatment method of Tomoeda et al. (1968) was also performed for plasmid curing. Loss of plasmid and antibiotic susceptibility testing of antibiotics to which bacteria were resistant were used to confirm the curing.
Statistic analysis
All analyses were carried out according to one-way analysis of variance (ANOVA) and assessed by post hoc comparison of means using lowest significant differences (LSD) using SPSS 11.0 software. They were considered significant at p < 0.05 level. The experiments were performed in triplicate.
Results
Isolation and identification of Escherichia spp
Six types of migrating wild birds and nine types of non-migrating birds were captured from Taif area for isolation of Escherichia spp. Captured birds were characterized and identified (Table 1). Pure colonies of bacteria were isolated from birds on MacConkey agar plates from cloacae samples and preliminary identification as Escherichia spp.
The distribution pattern of Escherichia spp. isolates is summarized in Table 2. The present results revealed that 92% of total samples collected from birds were found Escherichia spp. positive. The prevalence range of all two types of sample sources found positive was from 92% in non-migrating birds to 94% in migrating birds. Present study showed significantly a high percentage of migrating bird samples contained Escherichia spp. (p < 0.05).
Among bacterial strains isolated from birds, a total of 82 (50 isolates from non-migrating birds and 32 isolates from migrating birds) (Table 2) were selected and subjected to morphological and biochemical tests. The biochemical tests which were used for identification of Escherichia sp. isolates from birds are summarized in Table 3. The biochemical reactions revealed that all bacterial isolates belonged to two species of the genus Escherichia which were E. coli and E. vulneris. Also, API 20E identification confirmed the results obtained by the biochemical tests.
Virulence markers of Escherichia spp
To investigate the pathogencity of Escherichia spp. isolates, congo red binding tests were carried out. 8% of isolates obtained from non-migrating birds were strongly positive congo red binding whereas 25% of isolates from migrating birds were strongly positive (Table 4). These results indicated that positive congo red binding of isolates was significantly higher in migrating birds and lower in non-migrating birds (p < 0.05). Congo red binding has been used as a potential virulence marker, which indicated that these isolates were pathogens.
Moreover, in order to investigate hemolysin production prevalence among the bacterial isolates, hemolysis tests were achieved. 8% bacterial isolates obtained from non-migrating birds caused hemolysis in sheep blood agar plates while 75% of isolates from migrating birds caused hemolysis (Table 4). These results indicated that hemolysis of isolates was significantly higher in migrating birds and lower in non-migrating birds (p < 0.05).
Antimicrobial susceptibility of Escherichia sp isolates
Antimicrobial susceptibility profiles of 50 Escherichia spp. isolates from samples of non-migrating bird sources have been demonstrated in Table 5. Resistance spectrum of Escherichia spp. isolates for 15 antimicrobial agents tested in descending order was respectively Oxacillin, Lincomycin, Oxytetracycline, Amoxicillin, Cephalexin, Neomycin, Ampicillin, Cefaclor, Colistin, Piperacillin, Chloramphenicol, Ciprofloxacin, Norfloxacin, Gentamycin and Enrofloxacin. All isolates were found resistant to Oxacillin. 88%, 84% and 72% of isolates were found resistant to Lincomycin, Oxytetracycline and Amoxicillin, respectively. Moreover, 4% strains were found intermediate resistant to Cefaclor, Chloramphenicol, Gentamycin, Amoxicillin and Enrofloxacin, and 8% isolates were found intermediate resistant to Colistin, Oxytetracycline and Piperacillin. 32% and 48% isolates were found intermediate resistant to Neomycin and Cephalexin, respectively. 60-100% of isolates demonstrated sensitive to Cefaclor, Ampicillin, Chloramphenicol, Colistin, Ciprofloxacin, Norfloxacin, Gentamycin, Enrofloxacin and Piperacillin. Sensitive to Cefaclor, Ampicillin, Chloramphenicol, Colistin, Ciprofloxacin, Norfloxacin, Gentamycin, Enrofloxacin and Piperacillin was significantly high in the isolates obtained from non-migrating birds (p < .05). 72-100% of isolates demonstrated resistance to Amoxicillin, Oxytetracycline, Lincomycin and Oxacillin. Resistance to Amoxicillin, Oxytetracycline, Lincomycin and Oxacillin was significantly high in the isolates from non-migrating birds (p < 0.05).
Antimicrobial susceptibility profiles of 32 bacterial isolates of Escherichia spp. from samples of migrating birds have been presented in Table 6. Resistance spectrum of Escherichia spp. isolates for 15 antimicrobial agents tested in descending order was respectively Oxacillin, Chloramphenicol, Oxytetracycline, Lincomycin, Ciprofloxacin, Ampicillin, Cefaclor, Cephalexin, Amoxicillin, Colistin and Piperacillin. 100% of isolates were found resistant to Oxacillin Chloramphenicol, Oxytetracycline and Lincomycin. 87.5%, 75%, 50%, 37.5% and 12.5% of isolates were found resistant to Ciprofloxacin, Ampicillin, Amoxicillin, Colistin and Piperacillin, respectively. 62.5% of isolates were found resistant to Cefaclor and Cephalexin. Moreover, 6% and 94% strains were found intermediate resistant to Ampicillin and Neomycin, respectively, and 12.5% isolates were found intermediate resistant to Ciprofloxacin and Amoxicillin. 62.5-100% of isolates found sensitive to Colistin, Norfloxacin, Gentamycin, Enrofloxacin and Piperacillin indicative sensitive to these antimicrobial agents was significantly high in isolates of migrating birds (p < 0.05). 50-100% isolates demonstrated resistance to Oxacillin, Chloramphenicol, Oxytetracycline, Lincomycin, Ciprofloxacin, Ampicillin, Cefaclor, Cephalexin and Amoxicillin. These results indicated that resistance to Oxacillin, Chloramphenicol, Oxytetracycline, Lincomycin, Ciprofloxacin, Ampicillin, Cefaclor, Cephalexin and Amoxicillin was significantly high in isolates from migrating birds (p < 0.05).
MDR phenotype of Escherichia spp isolates
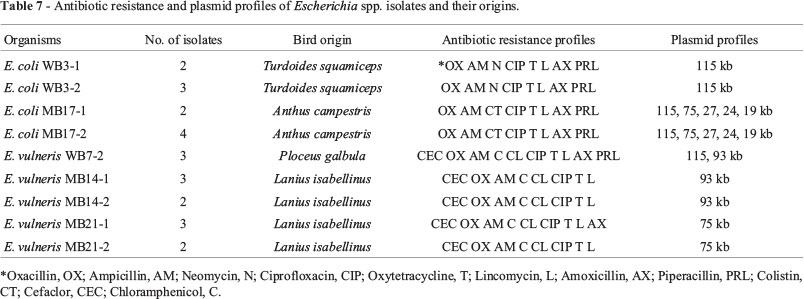
MDR patterns of Escherichia spp. isolates are shown in Table 7. Resistance to Oxacillin, Ampicillin, Neomycin, Ciprofloxacin, Oxytetracycline, Lincomycin, Amoxicillin and Piperacillin (OX,AM,N,CIP,T,L,AX,PRL) was significantly higher in the isolates from non-migrating bird, Arabian Babbler (Turdoides squamiceps), than in those from other non-migrating birds (p < 0.05). Isolates from non-migrating bird, Ruppell's Weaver (Ploceus galbula), were significantly more resistant to Cefaclor, Oxacillin, Ampicillin, Chloramphenicol, Cephalexin, Ciprofloxacin, Oxytetracycline, Lincomycin, Amoxicillin and Piperacillin (CEC,OX,AM,C,CL,CIP,T,L,AX,PRL) than those from other non-migrating birds (p < 0.05). Resistance to Cefaclor, Oxacillin, Ampicillin, Chloramphenicol, Cephalexin, Ciprofloxacin, Oxytetracycline, Lincomycin, Amoxicillin (CEC,OX,AM,C,CL,CIP,T,L,AX) was significantly higher in isolates from migrating bird, Isabelline Shrike (Lanius isabellinus) than in isolates from other migrating birds (p < 0.05).
Molecular characterization of most MDR Escherichia spp isolates
The bacterial isolates (WB3-1, WB3-2, MB17-1, MB17-2, WB7-2, MB14-1, MB14-2, MB21-1 and MB21-2) showed MDR phenotype were further identified by 16S rRNA encoding gene analysis. Part of the 16S rRNA encoding gene of these isolates was PCR-amplified and sequenced. The resulting nucleotide sequences were compared to available sequences in the databases. A phylogenetic tree illustrating the results of 16S rRNA analysis is demonstrated in Figure 1. As demonstrated, the 16S rRNA sequences of the isolates WB3-1, WB3-2, MB17-1 and MB17-2 are most closely related to E. coli (~98%) while as , the 16S rRNA sequences of the isolates WB7-2, MB14-1, MB14-2, MB21-1 and MB21-2 are most closely related to E. vulneris (~98%).
Plasmid profile
Agarose-gel electrophoresis of plasmid DNA from 9 isolates of MDR Escherichia (E. coli WB3-1, E. coli WB3-2, E. vulneris WB7-2, E. vulneris MB14-1, E. vulneris MB14-2, E. coli MB17-1, E. coli MB17-2, E. vulneris MB21-1 and E. vulneris MB21-2) is shown in Table 7 and Figure 2. Isolates E. coli WB3-1 and E. coli WB3-2 contain one large plasmid of ~115 kb whereas isolates E. coli MB17-1 and E. coli MB17-2 contain 5 plasmids of ~115 kb, 75 kb, 27 kb, 24 kb and 18 kb. Isolate E. vulneris WB7-2 contain two plasmid of ~115 kb and 93 kb. Isolates E. vulneris MB14-1 and E. vulneris MB14-2contain one plasmid of ~39 kb while as isolates E. vulneris MB21-1 and E. vulneris MB21-2 contain one plasmid of ~75 kb. The results showed that all of MDR Escherichia isolates contain a high-molecular weight plasmid DNA.
Plasmid curing studies of MDR Escherichia isolates were performed to detect if the drug resistance observed in this study was plasmid-mediated. No plasmid bands were detected after the electrophoretic separation of crude DNA extracts from cured isolates. Loss of plasmids correlated with loss of resistance to antibiotics in cured strains. Loss of resistance to multiple antibiotics was also demonstrated.
Discussion
Fifty isolates of Escherichia spp. recovered from non-migrating birds and 32 isolates from migrating birds were identified as two species of E. coli and E. vulneris. The prevalence range of samples found Escherichia spp. positive was from 92% in non-migrating birds to 94% in migrating birds. These results indicated significantly a high percentage of migrating bird samples contained Escherichia spp. Binding of congo red dye by E. coli is associated with the pathogenicity of the organism (Styles and Flammer, 1991). The present results indicated that positive congo red binding of Escherichia spp. isolates was significantly higher in migrating birds and lower in non-migrating birds. Congo red binding has been used as a potential virulence marker, which indicated that these isolates were pathogens. Moreover, hemolysin production is presumed to be a virulence factor in extraintestinal, e.g. urinary tract, infections caused by E. coli (Hacker, 1983). The results indicated that hemolysin production of isolates was significantly higher in migrating birds and lower in non-migrating birds. These results indicated that these isolates showed one to two virulence factors (congo red binding and hemolysis). E. coli is one of the common microbial flora of gastrointestinal tract of birds, animals and human but may become pathogenic to both (Levine, 1987). The present results indicated that migrating birds act as long-distance vectors for a wide range of Escherichia spp. that might be infectious to humans. This generates the potential for establishment of novel foci of emerging or re-emerging communicable diseases along bird migration routes.
On other hand, 100%, 88%, 84% and 72% of Escherichia spp. isolates from non-migrating birds were exhibited resistant to Oxacillin, Lincomycin, Oxytetracycline and Amoxicillin, respectively. Resistance to Amoxicillin, Oxytetracycline, Lincomycin and Oxacillin was significantly high in Escherichia spp. isolates from non-migrating birds. In addition, 50-100% of Escherichia spp. isolates recovered from migrating birds showed resistance to Oxacillin, Chloramphenicol, Oxytetracycline, Lincomycin, Ciprofloxacin, Ampicillin, Cefaclor, Cephalexin and Amoxicillin. Our results indicated that resistance to Oxacillin, Chloramphenicol, Oxytetracycline, Lincomycin, Ciprofloxacin, Ampicillin, Cefaclor, Cephalexin and Amoxicillin was significantly high in isolates from migrating birds. Previous results reported that 72% and 21% of E. coli isolates recovered from the faeces of wild Canada geese (Branta canadensis) and choughs (Pyrrhocorax pyrrhocorax) that were feeding and/or living in close to livestock waste were found resistant to amoxicillin-clavulanic acid of (Blanco et al., 2009; Cole et al., 2005). The wild birds in our study might have come into contact with antimicrobial residues and resistant bacteria from human waste in waterways. Moreover, because some antimicrobials utilized by human and veterinary medicine do not completely destroy, they may distribute through the environment in wastewater and soil (Sarmah et al., 2006). The difference in the prevalence of antimicrobial resistance in wildlife living in natural habitats in different geographic sites may reflect different levels of general pollution in the local environment (Osterblad et al., 2001). Wildlife, such as the black-headed gull (Larus ridibundus) or the Russian rook (Corvus frugilegus), might transport resistant bacteria from other areas to natural environments and then act as reservoirs, maintaining antimicrobial resistance within natural ecosystems (Blanco et al., 2009; Cole et al., 2005; Dolejska et al., 2007). A range of extended-spectrum β-lactamase-producing E. coli isolates recovered from seagull feces from Porto, Portugal, beaches exhibited a high rate of cefotaximase-15 resistance (Simoes et al., 2010).
All isolates from migrating and non-migrating birds examined in this study exhibited MDR phenotype to approximately 3-10 antimicrobial agents. Resistance to OX,AM,N,CIP,T,L,AX,PRL was significantly high in Escherichia spp. isolates recovered from non-migrating bird, Arabian Babbler (Turdoides squamiceps). Escherichia spp. Isolates recovered from non-migrating bird, Ruppell's Weaver (Ploceus galbula), were significantly more resistant to CEC,OX,AM,C,CL,CIP,T,L,AX,PRL. Resistance to CEC,OX,AM,C,CL,CIP,T,L,AX was significantly high in Escherichia spp. isolates recovered from migrating bird, Isabelline Shrike (Lanius isabellinus). MDR were significantly higher in the isolates from migrating birds than those from non-migrating birds. These results indicated that the isolates from non-migrating birds might gain the antimicrobial-resistance from isolates of migrating birds. Moreover, no significant differences were observed between the patterns of resistance among the swab sample isolates from either non-migrating birds or migrating birds indicative cross-transfer of antimicrobial-resistance. A high frequency of E. coli isolates from migrating Canada geese sampled on the eastern shore of Maryland in the United States were resistant to penicillin G, ampicillin, cephalothin, and sulfathiazole (Middleton and Ambrose, 2005). Due to indiscriminate exploitation of antimicrobial agents, such high prevalence of multi-drug resistance may apparently be occurred which may ultimately replace the drug sensitive microorganisms from antimicrobial saturated environment (Van de Boogard and Stobberingh, 2000). The present finding indicated that spreading of drug-resistant bacteria by migrating birds is worldwide. Wildlife is normally not clinically exposed to antimicrobial agents but can get antimicrobial resistant bacteria through contact with humans, domestic animals and the environment, and the water contaminated with faeces seems to be the most important vector.
Escherichia spp. isolates exhibited MDR phenotypes were further identified by 16S rRNA analysis. The results indicated greatest similarity to members of the Escherichia group, which matches the conclusions of the morphological, biochemical and API 20E analysis. The 16S rRNA gene of isolates WB3-1, WB3-2, MB17-1 and MB17-2 shares ~98% similarity with that of uropathogenic E. coli ATCC 25922 (American Type Culture Collection, Rockville, MD, USA) indicative that these bacteria are new isolates of the bacterium E. coli. The 16S rRNA gene of isolates WB7-2, MB14-1, MB14-2, MB21-1 and MB21-2 shares ~98% similarity with that of E. vulneris ATCC 33821. These results suggest that these bacteria are new isolates of the bacterium E. vulneris. Also, this is the first finding reported that E. coli and E. vulneris isolates originating from these birds carry MDA determinants. E. vulneris was recognized as a new species of the family Enterobacteriaceae only in 1982 and associated with human wounds (Brenner et al., 1982). E. vulneris has been isolated from animals, human, the environment, and potable water. E. vulneris can colonize the respiratory tract, female genital tract, urinary tract, and stool in human. The existence of several resistance to multiple classes of antimicrobial agents indicates that infection caused these isolates would be difficult to treat using antimicrobials currently available. The diversity of the cloacal microbial community in migrating and non-migrating birds, caught at Taif region, was assessed by cultivation and 16S rRNA analysis provide a better understanding of the bird' potential to harbor and disperse MDR pathogens.
Large plasmids of sizes ~ 115, 93, 75 kb were observed in MDR Escherichia isolates. These results indicated that plasmids with high molecular weights were found to be responsible for resistance to multiple antimicrobial compounds. Similar findings reported that large plasmids with sizes equal to or greater than 50 kb were observed in MDR E. coli isolates from animals (Karczmarczyk et al., 2011). Our results indicated that all of MDR Escherichia isolates contain a high-molecular weight plasmid DNA. High molecular weight plasmid found in MDR bacteria was reported by Lay et al. (2012). To detect the potential relation of multidrug resistance with plasmid in the bacterial isolates, plasmid curing was performed for the MDR Escherichia isolates. Curing of plasmid had resulted in loss plasmids. The plasmid cured cells became sensitive to all resistant antibiotics, indicative antibiotic resistance marker genes were located in plasmid. Our study conclude that multiple resistant Escherichia isolates and plasmid containing multidrug resistant genes are present in wild birds may act as a possible source of transfer of these highly resistant bacteria and their genes to human.
In conclusion, wild birds and especially migrating species can become long-distance vectors for a wide range of antimicrobial resistant microorganisms. Also, these results confirm the significant role of Escherichia spp. as reservoirs and carriers of multidrug-resistant plasmids. Therefore, more concerns are recommended for staff hygiene in processing and handling of birds and misuse of antimicrobial agents should be decreased or stopped by careful use of antimicrobial agents for the safety of public health. On the way to most successfully reduce the threat of diseases associated with wild birds, the public health and animal health sectors must work together in developing approaches to decline human contact to pathogens carried by non-migrating and migrating birds. A successful public educational operation could also put in viewpoint and clarify tradition and realities about the risk of obtaining infections associated with wild birds.
Acknowledgments
The authors do thank the Research Center of Taif University for funding this work through the research project No. 1-433-1845. We thank HH prince Bander Bin Saud, the head of Saudi Wildlife Authority (SWA), for his approval to capture the wild birds. Also, we are thankful to Prof. Dr. Simon C. Andrews, Reading University, England, for his assistant.
Submitted: April 4, 2013
Approved: April 17, 2014
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Altherr MR, Kasweck KL (1982) In situ studies with membrane diffusion chambers of antibiotic resistance and transfer in Escherichia coli Appl Environ Microbiol 44:838-843.
- Anderson DG, McKa LL (1983) Simple and Rapid Method for Isolating Large Plasmid DNA from Lactic Streptococci. Appl Environ Microbiol 46:549-552.
- Armstrong JL, Shigeno DS, Calomiris JJ, Seidler RJ (1981) Antibiotic-resistant bacteria in drinking water. Appl Environ Microbiol 42:277-283.
- Bell JB, Macrae WR, Elliott GE (1980) Incidence of R factors in coliform, fecal coliform, and Salmonella populations of the Red River in Canada. Appl Environ Microbiol 40:486-491.
- Blanco, G.; Lemus, J. A.; Grande, J. (2009) Microbial pollution in wildlife: linking agricultural manuring and bacterial antibiotic resistance in red-billed choughs. Environ. Res. 109, 405-412.
- Brenner DJ, McWhorter AC, Knutson JKL, Steigerwalt AG (1982) Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol 15:1133-1140.
- Buchanan RE, Gibbons NE (1974) Bergey's Manual of Determinative Bacteriology. 8th ed. Williams & Wilkins Co., Baltimore, Md.
- Calomiris JJ, Armstrong JL, Seidler RJ (1984) Association of metal tolerance with multiple antibiotic resistance of bacteria isolated from drinking water. Appl Environ Microbiol 47:1238-1242.
- Caprioli A, Donelli G, Falbo V, Passi C, Pagano A, Mantovani A (1991) Antimicrobial resistance and production of toxins in Escherichia coli strains from wild ruminants and the alpine marmot. J Wild Dis 27:324-327.
- Clesceri LS, Greenberg AE, Eaton AD (1998) Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, American Water Works Association, and Water Environment Federation, Washington, D.C.
- Clinical and Laboratory Standards Institute (CLSI) (2012) Performance standards for antimicrobial disk susceptibility tests. Approved standard M02-A11. National Committee for Clinical Laboratory Standards Institute. 11th ed. CLSI, Wayne.
- Cole D, Drum DJ, Stalknecht DE, White DG, Lee MD, Ayers S, Sobsey M, Maurer JJ (2005) Free-living Canada geese and antimicrobial resistance. Emerg Infect Dis 11:935-938.
- Dolejska M, Bierosova B, Kohoutova L, Literak I, Cizek A (2009) Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J Appl Microbiol 106:1941-1950.
- Dolejska M, Cizek A, Literak I (2007) High prevalence of antimicrobial resistant genes and integrons in Escherichia coli isolates from blackheaded gulls in the Czech Republic. J Appl Microbiol 103:11-9.
- Fortina MG, Silva M (1996) Mapping of two plasmids from Lactobacillus helveticus ILC54, plasmid curing and preliminary studies on their involvement in lactose metabolism and peptidase activity. Biotechnol Lett 18:1003-1006.
- Grobbel M, Lubke-Becker A, Alesik E, Schwarz S, Wallmann J, Werckenthin C, Wieler LH (2007) Antimicrobial susceptibility of Escherichia coli from swine, horses, dogs and cats as determined in the BfTGermVet monitoring program 2004-2006. Berl Munch Tierarztl Wochenschr 120:391-401.
- Hacker J, Schroter G, Schrettenbrunner A, Hughes C, Goebel W (1983) Hemolytic Escherichia coli strains in the human fecal flora as potential urinary pathogens. Zentralbl Bakteriol Mikrobiol Hyg A 254:370-378.
- Hubalek Z (1994) Pathogenic microorganisms associated with free-living birds (a review). Acta Scientiarum Naturalium Brno 28:1-74.
- Kanai H, Hashimoto H, Mitsuhashi S (1981) Drug resistance and conjugative R plasmids in Escherlchla coil strains isolated from wild birds (Japanese tree sparrows, green pheasants and bamboo partridges). Japanese Poultry Science 18:234-239.
- Karczmarczyk M, Abbott Y, Walsh C, Leonard N, Fanning S (2011) Characterization of multidrug-resistant Escherichia coli isolates from animals presenting at a University Veterinary Hospital. Appl Environ Microbiol 77:7104-7112.
- Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C (2009) Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol 75:559-566.
- Krumperman pH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165-170.
- Lay KK, Chansong N, Chuanchuen R (2012) Plasmid profiles of multidrug-resistant Escherichia coli from clinically healthy Swine. Thai J Vet Med 42:229-233.
- Levine M (1987) Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic and enteroadherent. J Infect Dis155:377-390.
- Martinez JL (2009) The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci 276:2521-2530.
- Middleton JH, Ambrose A (2005) Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J Wild Dis 41:334-341.
- Moffie BG, Mouton RP (1988) Sensitivity and resistance of Legionella pneumophila to some antibiotics and combinations of antibiotics. J Antimicrob Chemother 22:457-462.
- Murray GE, Tobin RS, Junkins B, Kushner DJ (1984) Effect of chlorination on antibiotic resistance profiles of sewage-related bacteria. Appl Environ Microbiol 48:73-77.
- Osterblad M, Norrdahl K, Korpimaki E, Huovinen P (2001) How wild are wild animals? Nature 409:37-38.
- Porter RF, Aspinall S (2010) Birds of the Middle East. Helm Field Guides. Cambridge.
- Sarmah AK, Meyer MT, Boxall AB (2006) A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725-759.
- Simoes RR, Poirel L, Da Costa PM, Nordmann . (2010) Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli Emerg Infect Dis 16:110-112.
- Sjolund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, Kahlmeter G, Olsen B (2008) Dissemination of Multidrug-Resistant Bacteria into the Arctic. Emerg Infect Dis 14:70-72.
- Styles DK, Flammer K (1991) Congo red binding of Escherichia coli isolated from the Cloacae of Psittacine birds. Avian Dis 35:46-48.
- Timoney JF, Port J, Giles J, Spanier J (1978) Heavy-metal and antibiotic resistance in the bacterial flora of sediments of New York Bight. Appl Environ Microbiol 36:465-472.
- Tomoeda M, Inuzuka M, Kudo N, Kakamura S (1968) Effective elimination of drug resistance and sex factors in Escherichia coli by sodium dodecyl sulphate. J Bacteriol 95:1078-1089.
- Trevors JT (1987) Survival of Escherichia coli donor, recipient, and transconjugant ceils in soil. Water Air Soil Pollut 34:409-414.
- Van de Boogard A E, Stobberingh EE (2000) Epidemiology of resistance to antibiotics links between animals and humans. Int J Antimicrob Agents 14:327-335.
- Walter MV, Vennes JW (1985) Occurrence of multiple-antibiotic resistant enteric bacteria in domestic sewage and oxidation lagoons. Appl Environ Microbiol 50:930-933.
- Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV (2001) Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother 45:2716-2722.
- Wnorowski AU (1993) Resistance to antibiotics of heavymetal tolerant and heavy-metal sensitive bacterial strains. J Environ Sci Health A28:203-215.
Publication Dates
-
Publication in this collection
19 Feb 2015 -
Date of issue
Dec 2014
History
-
Received
04 Apr 2013 -
Accepted
17 Apr 2014