Abstract
A mesophilic Enterobacter sp. Bn12 producing an alkaline thermostable lipase was isolated from soil in Tehran, Iran. The lipase gene (ELBn12) was identified from a genomic library. Sequence analysis of the DNA fragment revealed an open reading frame of 879 bp encoding a lipase with a molecular mass of 31.3 kDa. The deduced amino acid sequence showed 96% identity with a lipase of Enterobacter sp. Ag1 and the identity of their DNA sequences was 88.9%. ELBn12 belongs to the lipase subfamily I.1 and its catalytic triad consists of Ser82, Asp237 and His259. The lipase was expressed in Escherichia coli (BL21) pLysS and partially purified by anion exchange chromatography. The maximum activity of ELBn12 was obtained at temperature of 60 °C and pH 8.0 towards tricaprylin (C8) and its specific activity was around 2900 U/mg. ELBn12 was stable within a broad pH range from 6.0 to 11.0. The enzyme showed high stability in both polar and nonpolar organic solvents at 50% (v/v). The lipase activity was enhanced in the presence of 10 mM of Ca2+, Mg2+ and K+, while heavy metals (Fe3+ and Zn2+) had strong inhibitory effect. ELBn12 showed high activity in the presence of 1% (w/v) nonionic surfactants, however ionic surfactants inhibited the lipolytic activity. ELBn12 characteristics show that it has a potential to be used in various industrial processes.
Enterobacter; thermostable lipase; alkaline; organic solvent
RESEARCH PAPER
Cloning and characterization of newly isolated lipase from Enterobacter sp. Bn12
Parisa FarrokhI; Bagher YakhchaliII; Ali Asghar KarkhaneII
IDepartment of Genetics, School of Biological Science, Tarbiat Modares University, Tehran, Iran
IIIndustrial and Environmental Biotechnology Department, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
Correspondence Correspondence: B. Yakhchali Industrial and Environmental Biotechnology Department National Institute of Genetic Engineering and Biotechnology P.O. Box 14965/161, Tehran, Iran E-mail: bahar@nigeb.ac.ir
ABSTRACT
A mesophilic Enterobacter sp. Bn12 producing an alkaline thermostable lipase was isolated from soil in Tehran, Iran. The lipase gene (ELBn12) was identified from a genomic library. Sequence analysis of the DNA fragment revealed an open reading frame of 879 bp encoding a lipase with a molecular mass of 31.3 kDa. The deduced amino acid sequence showed 96% identity with a lipase of Enterobacter sp. Ag1 and the identity of their DNA sequences was 88.9%. ELBn12 belongs to the lipase subfamily I.1 and its catalytic triad consists of Ser82, Asp237 and His259. The lipase was expressed in Escherichia coli (BL21) pLysS and partially purified by anion exchange chromatography. The maximum activity of ELBn12 was obtained at temperature of 60 °C and pH 8.0 towards tricaprylin (C8) and its specific activity was around 2900 U/mg. ELBn12 was stable within a broad pH range from 6.0 to 11.0. The enzyme showed high stability in both polar and nonpolar organic solvents at 50% (v/v). The lipase activity was enhanced in the presence of 10 mM of Ca2+, Mg2+ and K+, while heavy metals (Fe3+ and Zn2+) had strong inhibitory effect. ELBn12 showed high activity in the presence of 1% (w/v) nonionic surfactants, however ionic surfactants inhibited the lipolytic activity. ELBn12 characteristics show that it has a potential to be used in various industrial processes.
Key words: Enterobacter, thermostable lipase, alkaline, organic solvent.
Introduction
Lipases (EC 3.1.1.3) belong to the family of carboxylic hydrolases, which catalyze both the hydrolysis and synthesis of esters (Treichel et al., 2010). They have common α/β fold and their active site usually consist of serin, histidin and aspartic acid (Arpigny and Jaeger, 1999). Lipases are an important group of biocatalysts for a wide variety of industrial applications (food, detergent and pharmaceutical industries) and account for about 5 to 10% of the biggest worldwide enzyme market (Salihu and Alam, 2012). Lipases have been isolated from plants, animals and microorganisms (Treichel et al., 2010). Microbial enzymes, because of their high production ability, ease of genetic manipulation and versatile catalytic activities in various environmental conditions, have greater advantages in industrial usage than the other sources (Andualema and Gessesse, 2012). Some of the desired characteristics for industrial application of lipases are stability in high temperatures, alkaline conditions and in the presence of organic solvents.
As the lipid substrates have high melting points, thermal stability is so crucial for lipases as biocatalysts in many industrial processes. Moreover thermophilic lipases show higher resistance to chemical denaturation (Lee et al., 1999). Stability of lipases in an organic solvent environment is beneficial since these solvents are able to increase the solubility of substrate, facilitate the recovery of nonpolar products, reduce the degree of undesirable substrate and/or product inhibition in organic solvent-water two-phase system (Rahman et al., 2003).
Thermostable lipases have potential applications in detergent, biodiesel, pharmaceutical and leather industries. Usage of organic solvent stable lipases has advantages in synthesis of biopolymers, biodiesel and pharmaceutical industry. Alkaline lipases are also being used for detergent, leather and pulp industries (Hasan et al., 2006).
With an increasing interest in biotechnological applications of lipolytic enzymes, searching on lipase-producing bacteria having satisfactory properties is an ongoing process. Although there are some characterized alkaline thermostable lipases from different microorganisms or metagenomic libraries (Ahmed et al., 2010; Guanasekaran et al., 2006; Khoramnia et al., 2010; Kim et al., 1996; Nawani et al., 1998; Sarkar et al., 2012), a few of them are stable in the presence of organic solvents (Ahmed et al., 2010; Guanasekaran et al., 2006; Nawani et al., 1998). In this study, we report functional screening, sequencing and characterization of a novel organic solvent- and thermo-stable alkaline lipase from a mesophilic Enterobacter species.
Materials and Methods
Bacterial strains and plasmids
The pTZ57R/T (Fermentas, Vilnius, Lithuania) and pBC KS+ (Agilent Technologies, Santa Clara, California, United States) were used for the PCR product cloning and genomic library construction respectively. Escherichia coli Top10 (Invitrogen, Grand Island, New York, United States) was used for cloning and plasmid preparation. E. coli (BL21) pLysS and pET-26(+) (Novagen, Madison, Wisconsin, United States) were used for expression of lipase gene.
Screening of thermostable alkaline lipase-producing bacteria
Two hundred fifty bacterial isolates, used in this study, were isolated from different soils (Zarenejad et al., 2012) and wastewaters of leather and edible oil industries (in this study) of Tehran, Iran. Lipase-positive colonies were detected by formation of clear halo on Luria-Bertani (LB) agar containing 0.5% (v/v) olive oil after 48 h of incubation at 37 °C. Among all isolates exhibiting lipolytic activity, isolates showing greater clear zones were subjected to hydrolysis of p-nitrophenyl palmitate (pNPP) (Sigma-Aldrich, Germany) at high temperature (50 °C) and alkaline conditions (pH range 9.0-10.0). Out of all isolates, one (Bn12) was selected for further studies because it showed the largest clear halo and superior hydrolysis activity at mentioned conditions.
Identification of the isolated bacterium
Genomic DNA was extracted from the isolate using phenol-chloroform method, as described by Sambrook et al. (2001). The 16S rRNA gene was amplified with two universal eubacterial primers: fD1 and rD1 (Weisburg et al., 1991). The PCR product was purified using an Agarose Gel DNA Purification Kit (Roche, Switzerland) and cloned into pTZ57R/T according to manufacturer's structure. DNA sequencing was carried out by dideoxy chain termination method (Macrogen, Seoul, Korea), using pUC/M13 primers.
The 16S rRNA gene sequence was analyzed through NCBI BLAST (http://www.ncbi.nlm.nih.gov/blast/) and EzTaxon servers (http://www.eztaxon.org/). The most similar sequences were aligned in Molecular Evolutionary Genetics Analysis (MEGA) 5.01 software and a phylogenetic tree was made by the neighbor-joining (NJ) method with 1,000 bootstrap replicates, in the MEGA (Tamura et al., 2011).
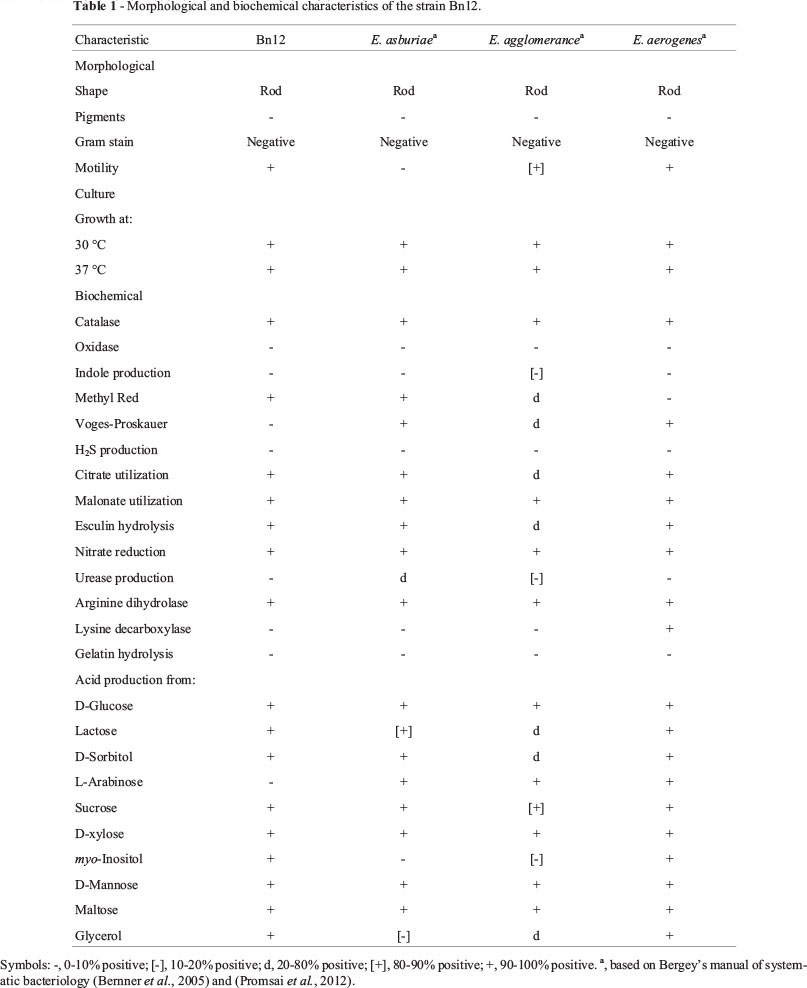
Morphological and biochemical identification of the isolate was made according to Bergey's manual of systematic bacteriology (Bernner et al., 2005).
Genomic DNA library construction
The chromosomal DNA was separately digested with Sau3AI, HindIII and pstI and then partial digest products with size of 1.5-10 kb were isolated by Agarose Gel DNA Purification Kit. These fragments were cloned into dephosphorylated BamHI-, HindIII- and pstI-digested pBC KS+ and transformed to E. coli TOP 10 by thermal shock transformation according to Sambrook et al. (2001). Screening of the transformants was carried out on 0.5% (v/v) olive oil LB agar plates supplemented with Chloramphenicol (30 μg/mL). Lipase-positive colonies were detected by the formation of clear haloes on olive oil LB agar plates after 48 h incubation at 37 °C.
Lipase sequence analysis
From a lipase-producing clone, the recombinant plasmid was extracted and sequenced with pUC/M13 primers. The upstream sequence of the lipase gene from genomic DNA of Bn12 strain was amplified using thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995) and then cloned and sequenced.
Homology searches against DNA and protein databases were performed using NCBI BLAST server. Multiple sequence alignment was done with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Theoretical molecular mass and isoelectric point value of the lipase were obtained using ExPASy bioinformatic resource portal (http://www.expasy.org/tools/). Prediction of N-terminal signal peptide sequence was done by SignalP 4.0 (Petersen et al., 2011) and SecretomeP 2.0 (Bendtsen et al., 2005) servers. Transmembrane regions analysis was performed using TMHMM 2.0 server (Krogh et al., 2001).
Cloning of the lipase gene
The lipase gene was amplified with the following primers; YKF1 (5'- ATCATATGTCTACATCCCTGAAGTAC-3') and YKF2 (5'- TAGAGCTCTTACAGTCCCTTCGCTTGC-3'). The underlined nucleotides indicate NdeI and SacI restriction sites respectively. The obtained PCR product was first cloned into the pTZ57R/T and then subcloned into the expression vector pET26(b+) using NdeI and SacI restriction sites. The nucleotide sequence of the insert in the resulting recombinant plasmid was confirmed by DNA sequencing. E. coli BL21 (DE3) pLysS was used for expression of recombinant lipase.
Expression and purification of the recombinant lipase
The E. coli BL21 (DE3) pLysS cells harboring the plasmid encoding lipase were grown in 200 mL LB medium supplemented with the required antibiotics at 37 °C until the absorbance at 600 nm of 0.6. The culture was then induced with a final concentration of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and incubated for another 18 h at 20 °C. After centrifugation, the bacterial pellet was resuspended in 20 mL of lysis buffer (50 mM Tris-HCl buffer (pH 8.5), 100 mM NaCl, 10 M EDTA, 1 mM PMSF, 1 mM DTT, 0.5% (v/v) Triton X-100) and disrupted by ultrasonic treatment (Hielscher GmbH, Teltow, Germany) at 4 °C.
The lysis extract was centrifuged at 15000 g for 15 min at 4 °C to pellet the cell debris and insoluble fractions. Supernatant was dialyzed overnight against 50 mM Tris-HCl buffer (pH 5.5). The dialyzed solution containing lipase was purified by anion exchange chromatography on diethylaminoethyl (DEAE)-cellulose column (1.5 x 7.0 cm) which had been equilibrated with the dialysis buffer. Under the experimental conditions used, most of the contaminating proteins bind to DEAE, while the lipase remains in unbounded protein fraction. Purity of the lipase was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970) and Coomassie Blue (G-250) staining and then dialyzed overnight against 20 mM Tris-HCl buffer (pH 8.0).
Zymographic analysis of lipase activity
Lipase activity was detected by zymography following standard SDS-PAGE. The SDS-PAGE gel was washed by agitating in 50 mM Tris-HCl buffer (pH 8.0) containing 2% (v/v) Triton X-100 for 1 h, and then rinsed 20 min with distilled water. The gel was placed on an agar plate containing 50 mM Tris-HCl buffer (pH 8.0), 0.5% (v/v) triglyceride and 20 mg/ml gum arabic (Sigma). Tributyrin and olive oil were used as substrates and appearance of hydrolysis halos exhibits lipase activity.
Protein estimation
The protein concentration of purified lipase was determined by Bradford's method using bovine serum albumin as the standard protein (Bradford, 1976).
Lipase assay
The lipase activity was measured titrimetrically using pH-Stat assay system (Metrohm Ltd., Herisau, Switzerland). Substrate solution comprised of 2% (w/v) gum arabic and 2 mg/ml triglycerides in deionized water. Emulsion of the solution was prepared by ultrasonic treatment. For lipase activity assay, 10 L of 0.2 mg/ml enzyme solution was added to the substrate solution and 0.1 M NaOH used as the titrant. One unit of lipase was determined as the amount of enzyme releasing 1 mol of fatty acid per min. All the experiments were done in triplicates.
Effect of temperature and pH on lipase activity and stability
Optimum temperature of the lipase activity was determined at different temperatures from 30 to 70 °C. The activity assay was carried out with tricaprylin (C8) at pH 8.0. Thermostability was assayed after incubation of lipase at 50, 60 and 70 °C for 1 h and the residual activity was analyzed every 20 min. Optimum pH for the lipase activity was measured over a pH range from 6.0 to 10.0 at 60 °C using tricaprylin as substrate. pH stability was determined at optimal conditions after incubating the lipase at 30 °C for 1 h over a pH value from 5.0 to 11.0. 50 mM sodium acetate was used for pH 5.0, 50 mM sodium phosphate buffer for pH 6.0-8.0 and 50 mM glycine-NaOH for pH 9.0-11.0.
Effect of substrate on lipase activity
Substrate specificity was studied by using triglyceride substrates; tributyrin (C4), tricaproin (C6), tricaprylin (C8), tricaprin (C10), trilaurin (C12), trimyristin (C14), tripalmitin (C16) and olive oil (C18) (Sigma) at 60 °C and pH 8.0.
Effect of various effectors on lipase activity
To determine the effect of various chemical effectors, the lipase activity was studied at optimal conditions after 1 h pre-incubating at 30 °C with 1% (w/v) of surfactants, 50% (v/v) of organic solvents, 10 mM of metal ions or 10 mM of EDTA.
Nucleotide sequence accession number
The nucleotide sequences of the 16S rDNA and ELBn12 lipase gene of Enterobacter sp. Bn12 were deposited in the EMBL/DDBJ/GenBank databases under the accession numbers JX456175 and JX456176 respectively.
Results
Isolation and identification of the lipase-producing bacterium
From 250 different isolates used in this study, 10 isolates with extracellular lipase activity were detected on LB agar containing 0.5% (v/v) olive oil through formation of clear haloes. Among these isolates, the Bn12 isolated from soil, with the largest clear zone and notable hydrolysis activity at alkaline pH and high temperature (data not shown), was selected for further studies.
On the basis of the most similar 16S rRNA sequences obtained from NCBI BLAST and EzTaxon servers, and also morphological and biochemical tests, the strain Bn12 was identified as Enterobacter sp. The results of morphological and biochemical traits have been summarized in Table 1. Based on the neighbor-joining tree (Figure 1), Bn12 strain occupies a distinct position within the genus Enterobacter.
Functional assay and analysis of the lipase gene and the flanking regions
From nearly 3500 E. coli transformants, one halo-forming colony was identified from pstI genomic library of Enterobacter sp. Bn12. Nucleotide sequencing of the recombinant plasmid, pLIP, from the halo-forming colony was shown a 1847 bp fragment. BLAST analysis of the fragment revealed that there was an ORF from position 89 to 967 related to lipases (Figure 2) which was named as ELBn12. Since the upstream region of the lipase gene was short, it was obtained through TAIL-PCR. Sequencing of the plasmid containing upstream region (pUPLIP) of the ELBn12 gene, revealed 881 bp fragment with 77% identity to a hypothetical protein (data not shown).
ELBn12 encodes a protein of 292 amino acids and BLASTx search of GenBank showed that the deduced ELBn12 had the most similarity to the putative lipases: 96% identity with lipase from Enterobacrter sp. Ag1 (EJF30243), and 70% identity with lipase from Yersinia enterocolitica (YP_004298016) over the entire length of the protein and 92% identity with LipC12 from metagenomic library (AEK97793) over 99% of the length of the protein. Identity of ELBn12 at the nucleotide level with the lipases of Enterobacrter sp. Ag1, LipC12, and Y. enterocolitica was 88.9% , 85.1% and 65.5% respectively.
There was a potential ribosome-binding site, GAGG, 10 bp upstream from the start codon. The putative -10 and -35 regions were predicted according to the E. coli promoter consensus sequences (McClure, 1985) (Figure 2).
Catalytic triad of the ELBn12 consists of serine (Ser82), aspartic acid (Asp237) and histidin (His259). The catalytic nucleophile Ser82 being present in Gly-His-Ser-Gln-Gly sequence of ELBn12 (Figure 3) matches with the conserved motif, Gly-X-Ser-X-Gly (where X is any amino acid), found in the family of lipases. The predicted molecular mass and pI of the ELBn12 were 31307.68 Da and 6.13 respectively. There were not any classical or non-classical predictable signal peptide and transmembrane domain in ELBn12 sequence.
Cloning, expression, and purification of ELBn12
The amplified ELBn12 (892 bp) by YKF primers was cloned into pET26(b+) and the resulting recombinant plasmid was named pYKF. The pYKF was transformed into E. coli BL21 (DE3) pLysS to express ELBn12 lipase. IPTG induction of pYKF at 20 °C for 18 h produced the maximum amount of ELBn12 lipase. The recombinant lipase was detected in the soluble fraction with molecular weight of approximately 30 kDa on SDS-PAGE which was close to the predicted size. The ELBn12 was purified in one step chromatography using DEAE-cellulose column. The purified lipase showed a single band on SDS-PAGE corresponding to molecular mass of ~30 kDa (Figure 4).
Zymogram analysis
Zymographic analysis showed that the purified lipase had lipolytic activity. The hydrolysis halos were detected in the proximity of the 30 kDa molecular weight marker, while clear zone for tributyrin substrate was larger than olive oil substrate (Figure 4).
Effect of temperature and pH on the lipase activity and stability
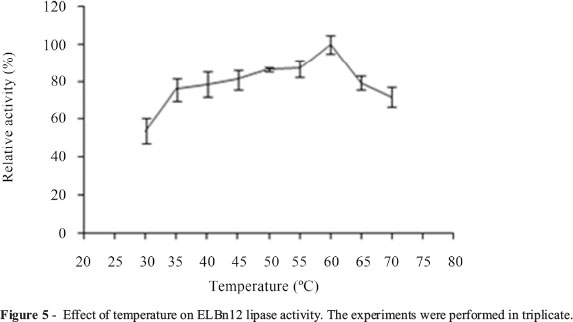
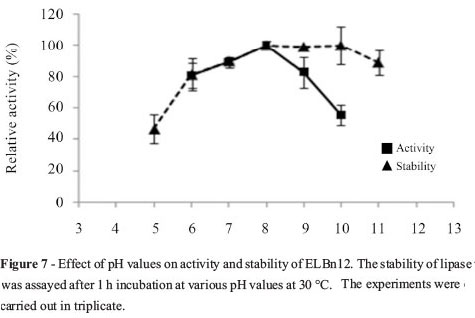
Purified ELBn12 exhibited maximum activity at temperature range of 45 to 65 °C with an optimum activity at 60 °C (Figure 5). ELBn12 retained more than 51% of its initial activity after 1 h incubation at 50 and 60 °C. However, the relative activity was reduced to 43% when incubated for 1 h at 70 °C (Figure 6). The ELBn12 showed the highest activity at pH 8.0. The enzyme retained more than 80% of its original activity after 1 h incubation at pH range from 6.0 to 11.0 (Figure 7).
Substrate specificity
The relative activity of the purified lipase against various triglycerides (C4 to C18) was determined at 60 °C; pH 8.0. ELBn12 did not show any specific activity for long or short chain triglycerides. It had a maximum activity towards C8 (100%), C12 (86.87%) and C4 (73.36%). The lipase activity was negligible against C16 and C10 (Figure 8). The specific activity of ELBn12 lipase was 2900 U/mg when measured at optimum conditions.
Effect of various effectors on the lipase activity
The lipase activity was partially activated by Ca2+, while K+, Mg2+ and Mn2+ increased enzyme activity slightly. Sodium ion had little inhibitory effect on the ELBn12 activity. In contrast, the enzyme activity was strongly inhibited by Zn2+ and Fe3+. The non-ionic detergents Tween 40, Triton X-100, Tween 80 and Tween 20 were found to increase (19 to 31%) the lipase activity. However, the ELBn12 activity was negligible in the presence of ionic detergents (SDS and cetyltrimethyl-ammonium bromide (CTAB)). Various organic solvents at 50% (v/v) partly enhanced the relative activity of the ELBn12 lipase. In the presence of 10 mM EDTA, the lipase was significantly stable (Table 2).
Discussion
At present study a novel organic solvent- and thermo-stable alkaline lipase was isolated from a mesophilic Enterobacter sp. Bn12 through genomic library. Although there are several studies describing cloning and characterization of microbial lipases (Chen et al., 2011; Kim et al., 1996; Rahman et al., 2003), a few of them describe characteristics of lipase from Enterobacter genus (Zhen-qian and Chun-yan, 2009).
Most of the thermostable enzymes are produced by thermophilic microorganisms, but there are some reports describing thermostable lipases from mesophilic ones. Some examples of these include thermostable lipases from Bacillus subtilis EH 37 (Ahmed et al., 2010), citrobacter freundii (Guanasekaran et al., 2006), Staphylococcus xylosus (Khoramnia et al., 2010), Acinetobacter sp. (Khoramnia et al., 2011), and Proteus vulgaris K80 (Kim et al., 1996).
According to the classification of Arpigny and Jaeger (1999), the primary structure of ELBn12 lipase and multiple sequence alignment of the most similar lipases showed that it was belonging to the subfamily I.1 of bacterial lipolytic enzymes. Molecular mass of ELBn12 was also in the range of the other members of subfamily I.1 (30-32 kDa) (Arpigny and Jaeger, 1999).
Similar to some lipases belong to the subfamily I.1 (Glogauer et al., 2011; Kim et al., 1996), there were no predictable signal sequences in the N- or C-terminal regions of ELBn12. Neither chaperon-encoding gene nor regulatory sequences were detected in the flanking regions of ELBn12. This observation, in addition to the functional expression of the lipase gene in E. coli BL21 (DE3) pLysS, indicates that its expression and folding do not depend on any helper proteins.
The maximum activity of ELBn12 was observed at 60 °C and pH 8.0 towards tricaprylin with specific activity of about 2900 U/mg. In comparison with the other thermostable bacterial lipases (Kim et al., 1996; Meilleur et al., 2009; Sarkar et al., 2012), ELB12 had notable specific activity. Unlike many of them (Khoramnia et al., 2010, 2011; Sarkar et al., 2012), the lipase showed less thermostability at high temperatures. Nevertheless, the specific activity of ELBn12 even after 1 h incubation at 70 °C (1000 U/mg) was remarkable and more than original activity of the many reported thermostable bacterial lipases (Khoramnia et al., 2011; Meilleur et al., 2009; Sarkar et al., 2012). ELBn12 also showed acceptable stability within a broad pH range from 6.0 to 11.0.
Stimulatory effects of some metal ions, such as Ca2+ were reported for several bacterial lipases (Andualema and Gessesse, 2012; Khan and Jithesh, 2012; Lee et al., 1999). Sharma et al. explained that this was due to the formation of insoluble Ca-salts of fatty acids released in the hydrolysis, thus avoiding product inhibition (2009). The result of this study showed that Ca2+ (10 mM) increased ELBn12 activity. However, its activity did not depend on the metal ions (Ca2+ and Mg2+) because in the present of chelating agent, EDTA (10 mM), the lipase maintained more that 90% of its original activity. Inhibitory effects of heavy metals (Fe3+ and Zn2+) on the lipase activity, as seen here, may be due to the alteration of enzyme conformation (Sharon et al., 1998).
Stability of lipases in different organic solvents was variable, but generally most of the lipases were more stable in the presence of nonpolar solvents than polar solvents (Ahmed et al., 2010; Rahman et al., 2003). In spite of destabilizing effect of polar solvents on the enzyme's conformation, the results of this study showed that both polar and nonpolar organic solvents at 50% (v/v) increased ELBn12 activity.
Catalytic activity of lipases is affected in the presence of surfactants differently. Many of the lipases were inhibited by ionic surfactants, while nonionic surfactants had stimulatory effects (Chen et al., 2011; Khoo and Ibrahim, 2009). The same result was observed for ELBn12.
Among the most similar lipases to the ELBn12, only LipC12 has been well characterized (Glogauer et al., 2011). In spite of their high amino acid sequence similarity (92%), the characteristics of ELBn12 were noticeably different from LipC12, especially in optimal temperature (maximum activity of LipC12 was at 30 °C). By characterization of more bacterial lipases, interpretation of these differences might be accessible in future. Furthermore, identification of new lipases with multiple appropriate characteristics has advantages to be applied in more industrial processes. ELBn12, which was described here, could be a useful candidate for application in various industries such as biodiesel, pharmaceutical and leather industries.
Acknowledgments
The authors would acknowledge the Najm Biotech Co Ltd for providing bacterial isolates from soil.
Submitted: February 11, 2013
Approved: September 9, 2013
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Ahmed EH, Raghavendra T, Madamwar D (2010) A thermostable alkaline lipase from a local isolate Bacillus subtilis EH 37: characterization, partial purification, and application in organic synthesis. Appl Biochem Biotechnol 160:2102-2113.
- Andualema B, Gessesse A (2012) Microbial lipases and their industrial applications: review. Biotechnol 11:100-118.
- Arpigny JL, Jaeger K (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177-183.
- Bendtsen JD, Kiemer L, Fausbøll A, Brunak S (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5:58.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248-254.
- Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) (2005) Bergey's manual of systematic bacteriology. Springer, New York.
- Chen R, Guo L, Dang H (2011) Gene cloning, expression and characterization of a cold-adapted lipase from a psychrophilic deep-sea bacterium Psychrobacter sp. C18. World J Microbiol Biotechnol 27:431-441.
- Glogauer A, Martini VP, Faoro H, Couto GH, Müller-Santos M, Monteiro RA, Mitchell DA, Souza EM, Pedrosa F, Krieger N (2011) Identification and characterization of a new true lipase isolated through metagenomic approach. Microb Cell Fact 10:54.
- Guanasekaran V, Kotay SM, Das D (2006) Alkaline lipase production by citrobacter freundii IIT-BT L139. Indian J Exp Biol 44:485-491.
- Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzyme Microb Technol 39:235-251.
- Khan M, Jithesh K (2012) Expression and purification of organic solvent stable lipase from soil metagenomic library. World J Microbiol Biotechnol 28:2417-24.
- Khoo ML, Ibrahim CO (2009) Lipase from thermoalkalophilic Pseudomonas species as an additive in potential laundry detergent formulations. Malays J Microbiol 5:1-5.
- Khoramnia A, Lai OM, Ebrahimpour A, Tanduba CJ, Voon TS, Mukhlis S (2010) Thermostable lipase from a newly isolated Staphylococcus xylosus starin; process optimization and characterization using RSM and ANN. Electron J Biotechnol 13:1-16.
- Khoramnia A, Ebrahimpour A, Beh BK, Lai OM (2011) Production of a solvent, detergent, and thermotolerant lipase by a newly isolated Acinetobacter sp. in submerged and soild-state fermentations. J Biomed Biotechnol 2011:702179.
- Kim HK, Lee JK, Kim H, Oh TK (1996) Characterization of an alkaline lipase from Proteus vulgaris K80 and the DNA sequence of the encoding gene. FEMS Microbiol Lett 135:117-121.
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567-80.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat 227:680-685.
- Lee DW, Koh YS, Kim KJ, Kim BC, Choi HJ, Kim DS, Suhartono MT, Pyun YR (1999) Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett 179:393-400.
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457-463.
- McClure WR (1985) Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54:171-204.
- Meilleur C, Hupé JF, Juteau P, Shareck F (2009) Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J Ind Microbiol Biotechnol 36:853-861.
- Nawani N, Dosanjh NS, Kaur J (1998) A novel thermostable lipase from a thermophilic Bacillus sp.: characterization and esterification studies. Biotechnol Lett 20:997-1000.
- Petersen TN, Brunak S, Heijne GV, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth 8:785-786.
- Promsai S, Tragoolpua Y, Jatisatienr A, Thongwai N (2012) Adhesion of wilt causing bacteria in Curcuma alismatifolia tissue. Int J Agric Biol 14:377-382.
- Rahman RNZA, Chin JH, Salleh AB (2003) Cloning and expression of a novel lipase gene from Bacillus sphaericus 205y. Mol Gen Genomics 269:252-260.
- Salihu A, Alam MZ (2012) Production and applications of microbial lipases: a review. Sci Res Essays 7:2667-2677.
- Sambrook J, Russell DW, (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, USA, 6.9-6.11 and 1.112-1.115.
- Sarkar P, Yamasaki S, Basak S, Bera A, Bag PK (2012) Purification and characterization of a new alkali-thermostable lipase from Staphylococcus aureus isolated from Arachis hypogaea rhizosphere. Process Biochem 47:858-866.
- Sharma A, Bardhan D, Patel R (2009) Optimization of physical parameters for lipase production from Arthrobacter sp. BGCC#490. Indian J Biochem Biophys 46:178-183.
- Sharon C, Furugoh S, Yamakido T, Ogawa H, Kato Y (1998) Purification and characterization of a lipase from Pseudomonas aeruginosa KKA-5 and its role in castor oil hydrolysis. J Ind Microbiol Biotechnol 20:304-307.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731-2739.
- Treichel H, Oliveira DD, Mazutti MA, Luccio MD, Oliveira JV (2010) A review on microbial lipases production. Food Bioprocess Technol 3:182-196.
- Weisburg W, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697-703.
- Zarenejad F, Yakhchali B, Rasooli I (2012) Evaluation of indigenous potent mushroom growth promoting bacteria (MGPB) on Agaricus bisporus production. World J Microbiol Biotechnol 28:99-104.
- Zhen-qian Z, Chun-yan G (2009) Screening for lipase-producing Enterobacter agglomerans for biodiesel catalyzation. Afr J Biotechnol 8:1273-1297.
Publication Dates
-
Publication in this collection
30 Sept 2014 -
Date of issue
June 2014
History
-
Accepted
09 Sept 2013 -
Received
11 Feb 2013