Abstract
This study reports the occurrence of antibiotic resistance and production of β-lactamases including extended spectrum beta-lactamases (ESβL) in enteric bacteria isolated from hospital wastewater. Among sixty-nine isolates, tested for antibiotic sensitivity, 73.9% strains were resistant to ampicillin followed by nalidixic acid (72.5%), penicillin (63.8%), co-trimoxazole (55.1%), norfloxacin (53.6%), methicillin (52.7%), cefuroxime (39.1%), cefotaxime (23.2%) and cefixime (20.3%). Resistance to streptomycin, chloramphenicol, nitrofurantoin, tetracycline, and doxycycline was recorded in less than 13% of the strains. The minimum inhibitory concentration (MIC) showed a high level of resistance (800-1600 µg/mL) to one or more antibiotics. Sixty three (91%) isolates produced β-lactamases as determined by rapid iodometric test. Multiple antibiotic resistances were noted in both among ESβL and non-ESβL producers. The β-lactamases hydrolyzed multiple substrates including penicillin (78.8% isolates), ampicillin (62.3%), cefodroxil (52.2%), cefotoxime (21.7%) and cefuroxime (18.8%). Fifteen isolates producing ESβLs were found multidrug resistant. Four ESβL producing isolates could transfer their R-plasmid to the recipient strain E. coli K-12 with conjugation frequency ranging from 7.0 x 10-3 to 8.8 x 10-4. The findings indicated that ESβL producing enteric bacteria are common in the waste water. Such isolates may disseminate the multiple antibiotic resistance traits among bacterial community through genetic exchange mechanisms and thus requires immediate attention.
beta-lactamase; ESβL; Antibiotic resistance; Enteric bacteria; plasmid transfer
MEDICAL MICROBIOLOGY
RESEARCH PAPER
Incidence and transferability of antibiotic resistance in the enteric bacteria isolated from hospital wastewater
Mohammad Zubair AlamI,# # Present address: King Fahd Medical Research Center, King Abdulaziz University, P.O. Box 80216, 21589 Jeddah, Saudi Arabia. All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC. ; Farrukh AqilI,* * Present address: Brown Cancer Center, Delia Baxter Research Building, Rm 320, University of Louisville, 40202 Louisville, KY USA. ; Iqbal AhmadI; Shamim AhmadII
IDepartment of Agricultural Microbiology, Aligarh Muslim University, Aligarh, India
IIDivision of Microbiology, Institute of Ophthalmology, JN Medical College and Hospital , Aligarh Muslim University, Aligarh, India
Correspondence Correspondence I. Ahmad Department of Agricultural Microbiology Aligarh Muslim University 202002 Aligarh, India E-mail: ahmadiqbal8@yahoo.co.in
ABSTRACT
This study reports the occurrence of antibiotic resistance and production of β-lactamases including extended spectrum beta-lactamases (ESβL) in enteric bacteria isolated from hospital wastewater. Among sixty-nine isolates, tested for antibiotic sensitivity, 73.9% strains were resistant to ampicillin followed by nalidixic acid (72.5%), penicillin (63.8%), co-trimoxazole (55.1%), norfloxacin (53.6%), methicillin (52.7%), cefuroxime (39.1%), cefotaxime (23.2%) and cefixime (20.3%). Resistance to streptomycin, chloramphenicol, nitrofurantoin, tetracycline, and doxycycline was recorded in less than 13% of the strains. The minimum inhibitory concentration (MIC) showed a high level of resistance (800-1600 µg/mL) to one or more antibiotics. Sixty three (91%) isolates produced β-lactamases as determined by rapid iodometric test. Multiple antibiotic resistances were noted in both among ESβL and non-ESβL producers. The β-lactamases hydrolyzed multiple substrates including penicillin (78.8% isolates), ampicillin (62.3%), cefodroxil (52.2%), cefotoxime (21.7%) and cefuroxime (18.8%). Fifteen isolates producing ESβLs were found multidrug resistant. Four ESβL producing isolates could transfer their R-plasmid to the recipient strain E. coli K-12 with conjugation frequency ranging from 7.0 x 10-3 to 8.8 x 10-4. The findings indicated that ESβL producing enteric bacteria are common in the waste water. Such isolates may disseminate the multiple antibiotic resistance traits among bacterial community through genetic exchange mechanisms and thus requires immediate attention.
Key words: beta-lactamase, ESβL, Antibiotic resistance, Enteric bacteria, plasmid transfer.
Introduction
Bacterial resistance to antimicrobial drugs, especially in nosocomial, commensal and zoonotic bacterial pathogens, has emerged as a global concern (Tanvir and Khiyani, 2011). As zoonotic agents, resistant bacteria within live-stock are proposed to be a significant risk to human health (Mathew et al., 2007). Fueling this concern is the wide-spread detection of multidrug resistant enteric pathogens. Antibiotics are intensively used in human, veterinary and agriculture and considered as the most important factor promoting the emergence, selection, and dissemination of resistant organisms (Catry et al., 2033; Houghton, 2002).
β-Lactamases are major determinants of resistance to β-lactam antibiotics. Coliform bacteria, isolated from human free environment, may induce a wide variety of resistant species by exposure to β-lactam drugs (Ash et al., 2002; Boon and Cattanach, 1999). Extended spectrum β-lactamase (ESβL) mediated resistance has been seen in the early years of 1980s in nosocomial enterobacteriaceae strains (Datta et al., 2004; Livemore, 1995).
The ESβLs are plasmid mediated enzymes and capable of hydrolyzing and inactivating a wide variety of β-lactams, including third generation cephalosporins and aztreonam. Most of these are derivatives of TEM and SHV β-lactamases (Bush, 1989). On the other hand, plasmid mediated β-lactamases showed resistance to α-methoxy β-lactam (cefoxitin, cefotetan) as well as oxyimino-β-lactams (cefotaxime, ceftriaxone and ceftazidime) (Papanicolaou et al., 1990; Rosenau et al., 2000). The plasmid mediated antibiotic resistance facilitate easy spread between species and most likely to occur in the gastrointestinal tract (Kruse and Sorum, 1994; Silva et al., 2006). Moreover, plasmids encoded ESβLs often showed co-resistance to aminoglycosides, sulfonamides and quinolones. Therefore infections caused by such strains may be difficult to treat due to diminish choices of antibiotics (Philippon et al., 2002).
Hospitals contain a concentrated reservoir of resistant coliform bacteria which are less common in community. The path between the hospital and the community runs both ways (Kunz and Brook, 2010). Therefore escape of such ESβL producing pathogenic strains from hospital environment to natural water bodies through wastewater discharge will be of much environmental and public health importance (Mathew et al., 2007). Lack of systematic study in this direction has prompted us to investigate the wastewater released from hospitals in the city of Aligarh, India. Wastewater discharge of JN Medical College and City Hospital are quantitatively tested both for E. coli and non E. coli enteric drug resistant bacteria and their potential to transfer R-plasmids to E. coli K-12.
Materials and Methods
Bacterial strains
Wastewater from hospital was collected during the winter period of 2004-05 and tested for viable enteric bacteria on selective plates (Salmonella-Shigella agar, Luria agar, EMB agar and MacConkey agar plates). Bacterial isolates obtained from selective plates were submitted to biochemical tests and results in sixty nine isolates representing different enteric bacteria. These isolates were differentiated into E. coli and non E. coli enteric bacteria. The latter group was identified presumptively, having bacteria from Shigella, Salmonella, Citrobacter, Enterobacter and Klebsiella, on the basis of standard biochemical tests as described by Cappuccino and Sherman (1995). These tests include Gram-staining, starch and gelatin hydrolysis, production of H2S and acids, indole, methyl red and Voges-Proskauer test, nitrate, citrate utilization, catalase and oxidase test and production of acid and gas from carbohydrates.
Identification
Bacterial isolates were selected on eosin methylene blue agar plates and colonies producing metallic shin were picked using sterile tooth-picks. The strains were further identified by biochemical tests as described by Cappuccino and Sherman (1995). E. coli isolates were confirmed serologically by Central Research Institute, Kasualy, India and grouped separately from non E. coli enteric bacteria in the present investigation. Non E. coli enteric bacteria were further identified tentatively as Shigella, Citrobacter and Klebsiella and those which cannot be identified were grouped as miscellaneous enteric bacteria including four Salmonella spp.
Determination of antibiotic susceptibility
Antibiotic discs/powders namely penicillin, ampicillin, cefuroxime, cefixime, cefotaxime, methicillin, nalidixic acid, cotrimoxazole, norfloxacin, doxycycline, tetracycline, nitrofurantoin, chloramphenicol, and streptomycin were purchased from Hi-Media Laboratory Ltd., Mumbai (India). Antibiotic sensitivity behavior of the isolated strains was determined by disc diffusion method as described by Bauer et al. (1996) as adopted by Ahmad et al. (1994).
Determination of minimum inhibitory concentration (MIC) of antibiotics
Minimum inhibitory concentrations were determined by agar spot method against test strains as per recommendation of WHO as adopted by Ahmad et al. (1994). The MIC was considered the lowest concentration of the antibiotic at which no visible growth of test strain occurred.
Assay for the detection of β-lactamase production
β-lactamase production in the test strains was detected by rapid iodometric method as described by Catlin (1975) and adopted by Ahmad and Aqil (2007). Briefly, the test strains were grown overnight on nutrient agar plates. Several colonies of the test strain were taken from the plates to make a dense suspension in 0.1 mL of penicillin solution (6 mg/ml) in a small test tube. After 30 min incubation at room temperature, 2 drops of starch solution and 1 drop of iodine reagent were added in reaction mixture leading to development of blue color due to starch-iodine reaction. The test tubes were shaken for 1 min at room temperature. Rapid decolorization of blue color indicated the presence of β-lactamase. No change in blue color for 10 min indicated negative results. Substrate specificity of β-lactamases was similarly tested against other β-lactam drugs (ampicillin, cloxacillin and cefixime) replacing penicillin-G in the test system. Strains of E. coli harboring transferable plasmids (E. coli MTCC 391 and E. coli 38) were used as positive controls.
Isolation of plasmid DNA and agarose gel electrophoresis
The plasmids were isolated from drug resistant enteric bacteria by the method of Kado and Liu (1981) and characterized by agarose gel electrophoresis (Maniatis et al., 1989). The molecular weight estimates of the isolated plasmids were determined by comparison of their relative mobilities on 1% agarose gels with standard DNA molecular weight markers as well as E. coli V517 plasmids as reference. The molecular weights were calculated by VersaDoc imaging system, model 5000 (Bio-Rad, Hercules, CA, USA).
Detection of ESβLs by DDST and on the basis of substrate specificity
The extended spectrum β-lactamases were detected by double disc synergy test (DDST) as described previously (Ahmad and Aqil, 2007). In the DDST, synergy was determined between augmentin (20 mg amoxycillin and 10 mg clavulanic acid) and a 30 mg disc of each third generation cephalosporins test antibiotic placed at a distance of 30 mm apart on Mueller-Hinton agar (Hi-Media Pvt. Ltd. Mumbai, India). The test organism was considered to produce ESβL, if the zone size around the test antibiotic disc increased towards the augmentin disc. This increase occurs because the clavulanic acid present in the augmentin disc inactivates the ESβL produced by the test organism. ESβL production was further confirmed by hydrolysis of β-lactams of first, second and third generations as described by Catlin (1975). β-Lactamases hydrolyzing third generation of β-lactam antibiotics in addition to first and second were designated as ESβLs producer.
Detection of transferable antibiotic resistance and β-lactamase production by conjugation
Drug resistance transfer studies were carried out using broth mating technique as described earlier (Ahmad et al., 1994). Briefly the donor and recipient (E. coli K-12) strains were grown in 5 mL nutrient broth, mixed in 1:2 ratios in 7 mL fresh nutrient broth and incubated overnight at 37 °C. Mix culture was spread evenly onto double antibiotics selection plates. Plates were incubated at 37 °C for overnight. Colonies grown onto double antibiotic plates were supposed to be transconjugants and tested for confirmation.
(a) Confirmation of transconjugant Colonies: Colonies from double antibiotic nutrient agar plates were transferred to double antibiotic MacConkey agar plates. Colorless colonies indicating non lactose fermenting were observed on the selection plates and confirmed as recipient E. coli K-12 Smr strains on the basis of their cultural, morphological and nutritional characters.
(b) Screening of transconjugants: Transconjugants observed on double antibiotic plates were further screened to possess antibiotic resistance markers and their ability to produce β-lactamases as described earlier.
(c) Percent of Transfer: Percent transfer of drug resistant marker was calculated as follows:
Results and Discussion
Indiscriminate use of antibiotics has led to an increasing incidence of antibiotic resistance among strains of enterobacteriaceae, worldwide (French, 2010; Hiramatsu et al., 1997). In the last two decades, ESβLs have emerged as a major contributor of drug resistance. In this study a total of 69 isolates of enteric bacteria were isolated from 50 samples of hospital wastewater in the vicinity of Aligarh city, India. On the basis of morphological, cultural and biochemical characteristics, these isolates were grouped into four different genera, Escherichia (18 isolates), Citrobacter (07), Shigella (11), and Klebsiella (04). Other twenty nine isolates could not be differentiated on the basis of biochemical tests and grouped as unidentified (miscellaneous) enteric bacteria. These characteristics of enteric bacteria were similar to those reported in literature (Ananthnarayan and Paniker, 1986). Further molecular characterization of these isolates is needed to explore their genetic linkages. The findings revealed that the wastewater discharged from hospital is heavily loaded with multiple drug resistant enteric bacteria and can be a plausible source for contamination to the other natural water bodies.
The multidrug resistant strains are common in the random population screened. The acquisition of antimicrobial resistance provides a mechanism for survival to bacterium in an otherwise hostile environment. Resistance is generally plasmid encoded or transposons mediated (Mather et al., 2007; Carattoli, 2003). These plasmids are free to mutate without lethal effects on the host cell. They can also move between species and even genera, accumulating genes by conjugation or recombination. Transfer of resistance R-factors to other members of the enterobacteriaceae especially E. coli, Salmonella, and Shigella etc. makes the problem an even greater concern to clinicians, particularly in controlling diseases like typhoid, dysentery, diarrhoea and other gastro-intestinal infections both in humans and animals (Ahmad et al., 1994; Silva et al., 2006).
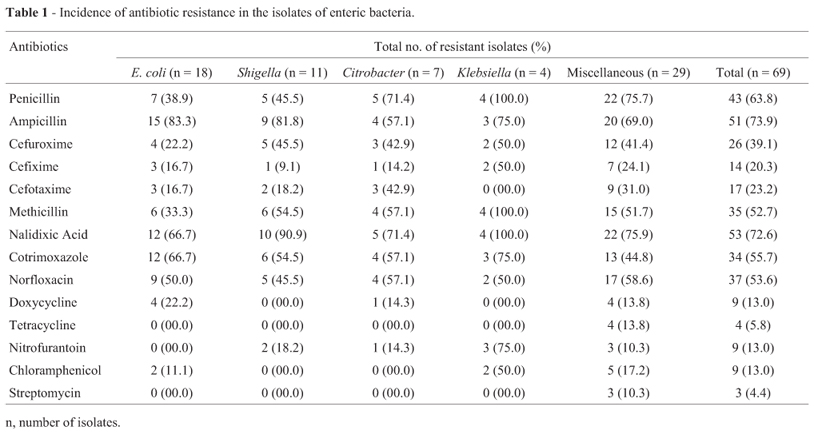
The resistance patterns against 14 antibiotics tested are presented in Table 1. Majority of the isolates showed resistance to one or more antibiotics. Such multiple antibiotic resistances is known to arise by acquisition of resistance genes through genetic exchange and mutation and physiological mechanisms such as possession of specific proteins and efflux pump (Hermansson et al., 1987; Herwig et al., 1997). Based on the disc diffusion test, incidence of antibiotic resistance was found maximum (73.9%) to ampicillin (A), followed by nalidixic acid (Na), penicillin (Pn), co-trimoxazole (Co), norfloxacin (Nx), methicillin (M), cefuroxime (Ce), cefotaxime (Cef), cefixime (Cfx), doxycycline (Do), nitrofurantoin (Nf), chloramphenicol (C), tetracycline (Tc) and least (4.4%) to streptomycin (Sm). Bacteria were more sensitive to cefuroxime and cefotaxime among the β-lactams. Similarly, less resistance was observed against protein synthesis inhibiting antibiotics such as tetracycline, doxycycline, chloramphenicol and streptomycin.
The resistance profile might be the results of excessive use of antibiotics in the treatment of bacterial infections. It is expected that these isolates have received antibiotic pressure in hospital environment during treatment. High resistance to β-lactams among enteric bacteria from clinical and environmental origin has been reported world-wide (Ash et al., 2002; Jain and Mondal, 2007; Rollinson, 1989). Resistance to nalidixic acid among clinical isolates of Pseudomonas and Staphylococcus is a common problem, but high incidence against enteric bacteria is less frequently reported (Murray, 1986; Rao et al., 2010).
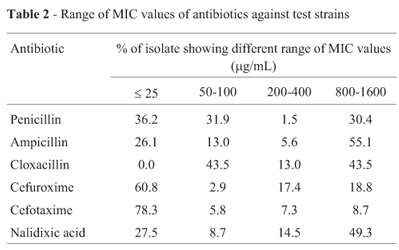
Increased incidence of ampicillin resistance was observed in E. coli (83.3%) followed by Shigella Klebsiella and Citrobacter sp. While miscellaneous constitute about 69.0% ampicillin resistant isolates. Klebsiella and Shigella were relatively more resistant to nalidixic acid compared to E. coli. The MIC data revealed that maximum number of isolates had higher level of resistance against ampicillin followed by nalidixic acid, penicillin, cefuroxime and cefotaxime (Table 2). The higher level of MIC and multiple antibiotic resistances in the enteric bacteria indicated the involvement of R-plasmids as the bacterial chromosome could not bear such an extra load of additional antibiotic resistance genes (Ahmad et al., 1994, Livermore and Williams, 1982).
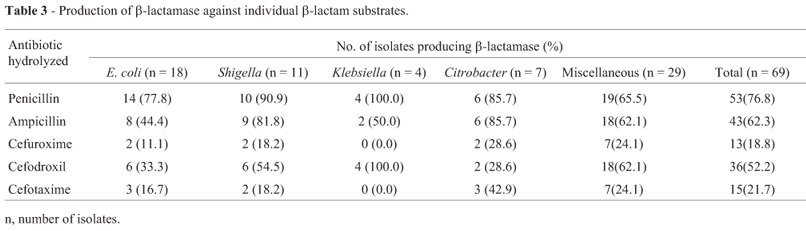
To ascertain the possible role of β-lactamases in the resistance against β-lactams, all the 69 isolates were tested for β-lactamase production using five substrates (penicillin, ampicillin, cefuroxime, cefodroxil, and cefotaxime). Of these, 63 isolates produced β-lactamase hydrolyzing one or more substrates (Table 3). β-Lactamases of these enteric bacteria hydrolyzed penicillin (76.8%) followed by ampicillin (62.3%), cefodroxil (52.2%), cefotaxime (21.7%) and cefuroxime (18.8%). The differential hydrolysis of β-lactam antibiotics by these enzymes is due to their substrate specificity (Bush, 1989). The data revealed that β-lactamase(s) production is the major mechanism of resistance against β-lactam antibiotics. Similar reports were also made by several workers among clinical isolates on enteric bacteria (Datta et al., 2004). However, environmental isolates of wastewater are less frequently screened for production of β-lactamases (Aubron et al., 2005).
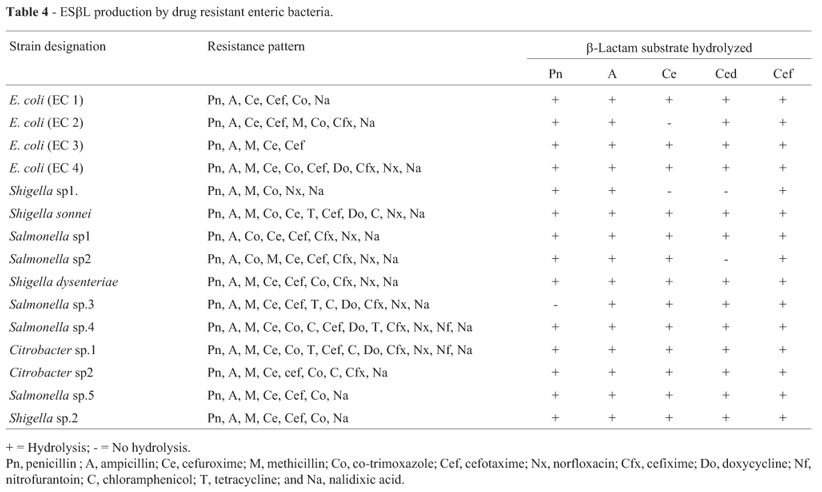
On the basis of hydrolysis of different generation of β-lactam antibiotics, 15 isolates of enteric bacteria were found to produce extended spectrum β-lactamases, a group of enzymes that confer resistance to oxyimino cephalosporins and monobactams (Jacoby and Medeiros, 1991). ESβLs are modification of the ubiquitous TEM and SHV class A enzymes, which have undergone a series of mutation at distinct sites in the enzyme that either increase the size of the active site or alter its binding characteristics to allow the larger cephalosporins to enter (Bush, 1989). The incidence of ESβL production was found maximum in the isolates of Citrobacter sp (42.86%) followed by the enteric bacteria of miscellaneous unidentified group including Salmonella spp. (24.14%), Shigella sp (18.18%) and E. coli (16.67%), while none of the Klebsiella isolate could produce ESβL (Table 4). Among clinical isolates of enteric bacteria ESβL production was reported with varying frequency (68-80%) by other workers (Mathur et al., 2002; Sahin et al., 2003). However, production of ESβL by enteric bacteria of environmental isolates is less frequently known.
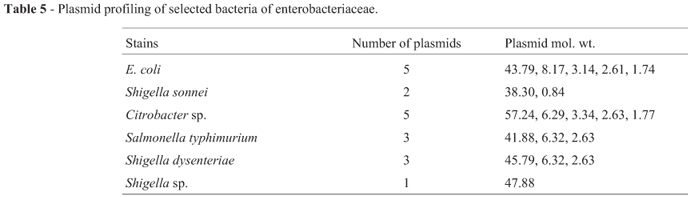
Plasmid profiling of these 15 multiple drug resistant isolates revealed the presence of more than one plasmid or the different forms of a single plasmid. The molecular weight of these plasmids ranged from few kb to 57.24 kb. Multiple bands with close molecular weight in a single bacterial strain may indicate the different forms of a single plasmid (Figure 1; Table 5). ESβL production among E. coli and other enteric bacteria are commonly encoded by plasmids (French, 2010; Livermore, 1982).
To investigate the transferable nature of these plasmids, all 15 ESβL producing strains were conjugated with E. coli K-12 (Smr recipient) strain. Interestingly, of these, 4 isolates could transfer ampicillin marker to the recipient with transfer frequency ranged from 2.6 x 10-4 to 1.8 x 10-5. In addition, two standard donor strains for β-lactamase, E. coli EC-38 and E. coli RP4 were also used as control. The frequency of conjugation of ampicillin marker for control strains ranged from 7.0 x 10-3 to 8.8 x 10-4 (Table 6). Our findings on transferable nature of β-lactamses encoding plasmids including ESBL are in agreement with the reports available in literature (Galani et al., 2002; Tait, 1993).
The finding of this investigation highlights the importance of safe disposal and better hospital wastewater treatment strategies to check the spread of drug resistant pathogenic bacteria in the environment as these may pose the public health on greater risk.
Submitted: November 24, 2011;
Approved: September 10, 2012.
- Ahmad I, Aqil F (2007) In vitro efficacy of bioactive extracts of 15 medicinal plants against ESβL producing multidrug resistant bacteria. Microbiol Res 162:264-275.
- Ahmad I, Yadava JNS, Ahmad S (1994) High level transferable resistance among E. coli Indian J Animal Sci 64:439-445.
- Ananthnarayan R, Paniker CKJ (1986) Enterobacteriaceae: Important distinguishing features of the different genera. A Text Book of Microbiology 3rd edition. Orient Longman Ltd Chennai. India.
- Ash RJ, Mauck B, Melissa-Morgan M (2002) Antibiotic resistance of Gram-negative bacteria in rivers, United States. Emerg Infect Dis 8:713-716.
- Aubron C, Poirel L, Ash RJ, Nordmann P (2005) Carbapene-mase-producing entero-bacteriaceae, US Rivers. Emerg Infect Dis 11:260-264.
- Baur AW, Kirby WMM, Sherris JC, Turch M (1966) Antibiotic susceptibility testing by a standardized single disc method. Amer J Clin Pathol 45:494-496.
- Boon PI, Cattanach M (1999) Antibiotic resistance of native and faecal bacteria isolated from rivers, reservoirs and sewage treatment facilities in Victoria, South-Eastern Australia. Lett Appl Microbiol 28:164-168.
- Bush K (1989) Characterization of β-lactamases. Antimicrob Agents Chemother 33:259-276.
- Cappuccino JG, Sherman N (1995) Microbiology: A Laboratory Manual. 3rd ed. The Benjamin/Cummings Pub Comp, Inc. Redwood City, California.
- Carattoli A (2003) Plasmid-Mediated Antimicrobial Resistance in Salmonella enterica Curr. Issues Mol Biol 5:113-122.
- Catlin BW (1975) Iodometric detection of Haemophilus influenzae β-lactamase: rapid presumptive test for ampicillin resistance. Antoimicrob Agent Chemother 7:265-270.
- Catry B, Laevens H, Devriese LA, Opsomer G, Kruif AD (2003) Antimicrobial resistance in livestock. J Vet Pharmacol Therap 26:81-93.
- Datta P, Thakur A, Mishra B, Gupta V (2004) Prevalence of clinical strains resistant to various beta lactams in a tertiary care hospital in India. Jap. J Infect Dis 57:146-149.
- French GL (2010). The continuing crisis in antibiotic resistance. Int J Antimicrob Agents 36:S3-S7.
- Galani I, Xirochauki E, Kanallakopoulou K, Petrikoss G, Giamarellou H (2002) Transferable plasmid mediating resistance to multiple antimicrobial agents in Klebsiella pneumoniae isolates in Greece. Clin Microbiol Infect 8:579-88.
- Hermansson M, Jones GW, Kjelleberg S (1987) Frequency of antibiotic and heavy metal resistance, pigmentation and plasmids in bacteria of the marine air-water interface. Appl Environ Microbiol 53:2338-2342.
- Herwig RP, Gray JP, Weston DP (1997) Antibacterial resistant bacteria in surficial sediments near salmon net-cage farms in Puget Sound, Washington. Aquaculture 149:263-283.
- Hiramatsu K, Aritaka N, Hanaki H (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus hetrogenously resistant to vancomycin. Lancet 324:601-612.
- Houghton D (2002) Antimicrobial resistance in the intensive care unit: understanding the problem. AACN Clin 3:410-420.
- Jacoby GA, Medeiros AA (1991) More extended Spectrum β-lactamases. Antimicrob Agents Chemother 35:1697-1704.
- Jain A, Mondal R (2007) Prevalence and antimicrobial resistance pattern of extended spectrum b-lactamase producing Klebsiella spp isolated from cases of neonatal septicaemia. Indian J Med Res 125:89-94.
- Kado CI, Liu SI (1981) Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365-1373.
- Kruse H, Sorum H (1994) Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl Environ Microbiol 60:4015-4021.
- Kunz AN, Brook I (2010) Emerging resistant Gram-negative aerobic bacilli in hospital-acquired infections. Chemother 56:492-500.
- Livemore DM (1995) β-lactamases in Laboratory and clinical resistance. Clin Microbiol Rev 8:557-584.
- Livermore DM, Williams RJ (1982) Current Chemotherapy and Immunotherapy. American Society for Microbiology. Washington DC.
- Maniatis T, Fritsch EF, Sambrook J (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbour, New York.
- Mathew AG, Cissell R, Liamthong S (2007) Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 4:115-33.
- Mathur P, Kapil A, Das B, Dhawan B (2002) Prevalence of extended spectrum beta lactamase producing Gram negative bacteria in a tertiary care hospital. Indian J Med Res 115:153-157.
- Murray BE (1986) Resistance of Shigella, Salmonella and other selected enteric pathogens to antimicrobial agents. Rev Infect Dis 8:S172-S181.
- Papanicolaou GA, Medeiros AA, Jacoby GA (1990) Novel plasmid mediated β-lactamase (MR-1) conferring resistance to oxyiminoand α-methoxy β-lactams in clinical isolates of K. pneumoniae. Antimicrob Agents Chemother 34:2200-2209.
- Philippon A, Arlet G, Jacoby GA (2002) Plasmid-determined AmpC-type beta lactamase. Antimicrob Agents Chemother 46:1-11.
- Rao S, Van-Donkersgoed J, Bohaychuk V, Besser T, Song XM, Wagner B, Hancock D, Renter D, Dargatz D, Morley PS (2010) Antimicrobial drug use and antimicrobial resistance in enteric bacteria among cattle from Alberta feedlots. Foodborne Pathog Dis 7:449-57.
- Rollinson GN (1989) β-lactamase induction and resistance to β-lactam antibiotics. J Antimicrob Chemother 23:1-5.
- Rosenau A, Cattier B, Gousset N, Harrian P, Phillippon A, Quentin R (2000) Capnocytophaga ochracea: characterization of a plasmid encoded expanded-spectrum TEM -17 beta lactamase in the phylum Flavobacter-Bacteroides. Antimicrob Agents Chemother 44:760-762.
- Sahin I, Kaya D, Oksuz S, Okay A, Sencan I, Ozturk E (2003) The frequency of extened spectrum beta lactamase and antibiotic susceptibility in clinical isolates of gram negative bacilli. Turkish J Infect 17:145-148.
- Silva J, Castillo G, Callejas L, López H, Olmos J (2006) Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electronic J Biotechnol. 9:533-540.
- Tanvir F, Khiyani F (2011) Antibiotic resistance: a global concern. J Coll Physicians Surg Pak. 21:127-129.
- Tait S (1993) Mobile genetic elements in antibiotic resistance. J Med Microbiol 38:157-159.
Publication Dates
-
Publication in this collection
03 Feb 2014 -
Date of issue
Sept 2013
History
-
Received
24 Nov 2011 -
Accepted
10 Sept 2012