Abstract
The petroleum-derived degrading Dietzia cinnamea strain P4 recently had its genome sequenced and annotated. This allowed employing the data on genes that are involved in the degradation of n-alkanes. To examine the physiological behavior of strain P4 in the presence of n-alkanes, the strain was grown under varying conditions of pH and temperature. D. cinnamea P4 was able to grow at pH 7.0-9.0 and at temperatures ranging from 35 ºC to 45 ºC. Experiments of gene expression by real-time quantitative RT-PCR throughout the complete growth cycle clearly indicated the induction of the regulatory gene alkU (TetR family) during early growth. During the logarithmic phase, a large increase in transcriptional levels of a lipid transporter gene was noted. Also, the expression of a gene that encodes the protein fused rubredoxin-alkane monooxygenase was enhanced. Both genes are probably under the influence of the AlkU regulator.
Dietzia cinnamea; n-hexadecane assimilation; alk genes; rubredoxin-alkane monooxygenase; real-time quantitative RT-PCR
ENVIRONMENTAL
RESEARCH PAPER
Transcriptional profiling of genes involved in n-hexadecane compounds assimilation in the hydrocarbon degrading Dietzia cinnamea P4 strain
Luciano ProcópioI, II; Michele de Cassia Pereira e SilvaII; Jan Dirk van ElsasII; Lucy SeldinI
IInstituto de Microbiologia Prof. Paulo de Góes, Universidade Federal do Rio de Janeiro, Centro de Ciências da Saúde, Ilha do Fundão, Rio de Janeiro, RJ, Brazil
IIDepartment of Microbial Ecology, University of Groningen, Kerklaan, Haren, The Netherlands
Send correspondence to Send correspondence to: L. Seldin Laboratório de Genética Microbiana, Departamento de Microbiologia Geral, Centro de Ciências da Saúde, Instituto de Microbiologia Prof. Paulo de Góes, Universidade Federal do Rio de Janeiro Bloco I, Ilha do Fundão 21941-590 Rio de Janeiro, RJ, Brazil E-mail: lseldin@micro.ufrj.br; lucy@seldin.com.br
ABSTRACT
The petroleum-derived degrading Dietzia cinnamea strain P4 recently had its genome sequenced and annotated. This allowed employing the data on genes that are involved in the degradation of n-alkanes. To examine the physiological behavior of strain P4 in the presence of n-alkanes, the strain was grown under varying conditions of pH and temperature. D. cinnamea P4 was able to grow at pH 7.0-9.0 and at temperatures ranging from 35 ºC to 45 ºC. Experiments of gene expression by real-time quantitative RT-PCR throughout the complete growth cycle clearly indicated the induction of the regulatory gene alkU (TetR family) during early growth. During the logarithmic phase, a large increase in transcriptional levels of a lipid transporter gene was noted. Also, the expression of a gene that encodes the protein fused rubredoxin-alkane monooxygenase was enhanced. Both genes are probably under the influence of the AlkU regulator.
Key words: Dietzia cinnamea, n-hexadecane assimilation, alk genes, rubredoxin-alkane monooxygenase, real-time quantitative RT-PCR.
Introduction
Petroleum-derived hydrocarbons cover a wide range of linear alkanes of different sizes, cycle-alkanes and aromatic hydrocarbons of natural origin. Microorganisms able to degrade such hydrocarbons have received greater attention in recent years. This is mainly due to their potential for bioremediation of oil spills or applications in biotransformations of fine chemicals (Atlas and Atlas, 1991). The utilization of aliphatic alkanes by bacteria is driven by two important steps: the transport of alkane compounds into the cell and a first reaction catalyzed by a monooxygenase, which converts alkanes to alkanols, also called hydroxylases (van Beilen and Witholt, 1994). These enzymes constitute a set of related non-heme iron integral membrane oxygenases, collectively named alkane hydroxylase (AlkB) (Smits et al., 1999). Currently there are three categories of alkane hydroxylases (van Beilen and Funhoff, 2007), i.e. methane monooxygenase, active on short-chain alkanes (C1-C4), membrane-bound non-heme iron and cytochrome P450 monooxygenases, which act on medium-chain alkanes (C5-C16) (van Beilen and Witholt, 1994) and poorly-characterized hydroxylases that oxidize long-chain alkanes (> C16) (Feng et al., 2007; Throne-Holst et al., 2007). The complete biochemical pathway responsible for assimilation of n-alkanes is most extensively described in Pseudomonas putida Gpo1 (van Beilen et al., 2001). In this species, the genes are arranged in the alkBFGHJKL operon localized in the OCT plasmid, containing all genes involved in hydrocarbon degradation (Van Hamme et al., 2003).
While there are several studies on the hydroxylation of n-alkanes by monooxygenases, the understanding of the acquisition of such hydrophobic compounds remains difficult. The low solubility of alkanes raises the question about the best strategies adopted by the cell for the acquisition of such compounds. Selective and active transport, which is energy-dependent, has been suggested by Beal and Betts (2000), who used an inhibitor of cytochrome oxidase to block oxidative phosphorylation, confirming the reduction in n-hexadecane uptake. A possible membrane lipoprotein (Blc) is suggested to be directly involved in the transport of alkanes to the inside of cells of Alcanivorax borkumensis (Sabirova et al., 2011). Another possible process involved in the acquisition of n-alkanes is the biosynthesis of biosurfactant, which apparently increases the solubility of n-alkanes through changes in its physical behavior (Van Hamme and Ward, 2001).
In recent years, at least 60 genera of aerobic bacteria and five anaerobic bacteria have been described to be able to degrade hydrocarbons of petroleum (Sakai et al., 1994; Prince, 2005). Among these bacteria, we find Rhodococcus (Van Hamme and Ward, 2001), Alcanivorax (Liu et al., 2010) and Pseudomonas (Zhang et al., 2011). Recently, species of Dietzia have emerged as potential degraders of petroleum-derived compounds. Dietzia spp. have been isolated from diverse environments, such as tropical soil (von der Weid et al., 2007), soda lakes (Duckworth et al., 1998), oil fields (Borzenkov et al., 2006), deep-sea sediments (Colquhoun et al., 1998), skin and intestinal tracts of marine fish (Yumoto et al., 2002) and decomposing reed rhizomes (Borsodi et al., 2005). In addition, studies of n-alkane degradation by Dietzia species have shown the ability to use different compounds (C6-C40) (Bihari et al., 2011; Wang et al., 2011). In a previous study of the effect of biostimulation on the diversity of the bacterial community present in a sandy loam (Cambisol) soil with no history of previous contamination, Dietzia cinnamea strain P4 was isolated (Evans et al., 2004). Subsequent studies of its biodegradation potential of petroleum-derived compounds revealed that strain P4 is able to grow on different sizes of linear and aromatic hydrocarbons (von der Weid et al., 2007).
Recently, the D. cinnamea P4 genome has become available in the NCBI database under the access number NZ_AEKG01000000. This genome is the first genome of a Dietzia species that has been sequenced and annotated. The genome annotation identified several enzymes of biotechnological interest and confirmed the presence of the genes involved in hydrocarbon compound degradation, e.g. n-alkanes, biphenyl and benzene compounds (Procópio et al., 2012). Here, we described the detailed genetic organization of these genes in the strain P4 chromosome, and the profile of degradation of n-hexadecane compounds, assessing the transcript levels of alk genes during growth by real-time RT-PCR.
Materials and Methods
Chemicals and oligonucleotide primers
Bushnell-Haas mineral salts medium (BH) was purchased from Difco, BD (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). n-Hexadecane (99% pure) and glucose were purchased from Sigma Co (St. Luis, MO, USA). Luria-Bertani Agar (LB-agar, tryptone 1%, NaCl 0.5%, yeast extract 0.5%, agar 1.5%) medium was obtained from Deutche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Germany. The primers used for the amplification of the interested genes are described in the Table 1.
Bacterial strain and growth conditions
All experiments were conducted with Dietzia cinnamea P4 strain in aerobic conditions. From a single colony, previously grown on LB-agar, a pre-inoculum was prepared in 50 mL Erlenmeyer flasks containing 10 mL of sterile BH medium (pH 7.0) supplemented with 1 g/liter of glucose (wt/vol) or 1% of n-hexadecane (vol/vol), at 28 ºC (under agitation -120 rpm) up to the late-exponential phase of growth. The n-hexadecane compound was filter-sterilized using a solvent-resistant Whatman PVDF sterile syringe filter. Then 1 mL of each pre-inoculum was transferred in 250 mL Erlenmeyer flasks containing 100 mL of BH medium supplement with 1 g/liter of glucose (wt/vol) or 1% of n-hexadecane (vol/vol), following the same conditions described above. In order to evaluate the growth in different pHs, the P4 strain was cultivated in BH medium at pH 5.0, 7.0 and 9.0, previously adjusted with HCl or NaOH and supplemented with 1% of n-hexadecane (vol/vol). The cultures were grown in three replicate flasks during 10 days. Sampling (1 mL) was done every day, centrifuged at 8,000g for 4 min, then the cell pellets were washed twice in 1 mL of sterile BH, and then optical density was measured at 660 nm to determine its growth curve.
Quantitative real-time PCR
One mL of each culture containing n-hexadecane as the sole carbon source was taken 2 hours after the onset of growth (early phase), in the late-exponential and mid-stationary phases, and growth in glucose as sole carbon source was taken only in late-exponential phase. Total RNA was extracted using the RNeasy Mini kit (Quiagen, Valencia, CA, USA) according to the manufactures protocol with the addition of one initial step of nitrogen liquid for cell lyses due to the difficulty of breaking the cell wall of P4 strain. Then the total RNAs were treated with DNase I (Promega, São Paulo, Brazil). The integrity of the RNAs was checked by agarose gel electrophoresis and the yield was estimated using a Nanodrop UV spectrometer (Thermo Scientific, Wilmingon, DE, USA). About 100 ng of RNA from each sample were used for cDNA synthesis using random hexamers by the cDNA Synthesis kit (Bioline, Boston, Ma, USA) according to the manufactures protocol. The yield of cDNAs was estimated by Nanodrop UV spectrometer and their concentrations adjusted to about 1 ng. For real-time quantitative RT-PCR, 2 µL of cDNA was mixed with Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA), 0.2 µg of forward and reverse primers of each gene to by analyzed (Table 1) in a final volume of 25 µL in three replicates. In addition, no-templates controls, in three replicates for each gene to by analyzed, also were included. Expression of the 16S rRNA gene (Table 1) was used as a reference gene to normalize tested genes and was correlated to the amount of corresponding transcripts in samples grown on glucose. The real-time quantitative RT-PCR reaction was carried out with the ABI Prism 700 Sequence Detection System (Applied Biosystems), following the protocol: one cycle at 95 ºC for 10 min, 40 cycles at 95 ºC for 1 min, followed by 60 ºC for 1 min, 72 ºC for 2 min. The specificity of the amplification was verified at the end of the PCR run through uses of the ABI Prism Dissociation Curve Analysis software, and the normalized relative fold change in mRNA levels were calculated for the gene of interest in each sample using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Sequence analysis
Sequence analysis of interesting genes and visualization of their organization in the strain P4 genome was performed using the Artemis program (Rutherford et al., 2000). Searches for conserved domains were performed using the BLAST suite (Altschul et al., 1990) and the transmembrane domains were identified using THMMH (Krogh et al., 2001). Alignments between the deduced amino acid sequences were performed by Clustal W (Thompson et al., 2002). Phylogenetic analysis of the analyzed genes were structured by maximum-parsimony method using the MEGA4 program (Tamura et al., 2007) and bootstrapping analysis was used to evaluate the tree topology by 1000 re-samplings.
Results
Genetic analysis of alk genes
The genome annotation of D. cinnamea P4 enabled the identification of a DNA region of 14.9 kbp, which contained 15 ORFs (access number: NZ_AEKG01000061), three of which are annotated as directly involved in the assimilation of aliphatic hydrocarbons (Procópio et al., 2012). ORF5 (ZP_08022272), with 1860 bp, was recorded initially as a putative ABC transporter permease. Subsequent analysis of similarity indicated 99% of homology of this gene with the putative ABC transporter of Dietzia sp. E1. The program THMMH identified five transmembrane domains. Next, ORF6, annotated as an alkane 1-monooxygenase (ZP_08022271), with 1527 bp, revealed upon subsequent analysis 98% of homology with an alkane hydroxylase/rubredoxin fusion protein. This protein contains six transmembrane domains. ORF7 was annotated as a puta-tive TetR family transcriptional regulator (ZP_08022270), with 625 bp. The probable arrangement of these genes on the chromosome of strain P4, with the presence of alkB-rub natural fusion genes, is very similar to that previously described in the related Dietzia sp. E1 (Figure 1). The putative alkane monooxygenase hydroxylase gene (alkB-rub, ORF6) is arranged in fusion with a rubredoxin gene. Thus, the probable alkB-rub gene is flanked by one TetR family transcriptional regulator (ORF7), which is described as alkU, in other degrader bacteria, and a putative bifunctional ABC lipid transporter (ORF5).
Multiple alignments between the full-length sequences of D. cinnamea P4 (ZP_08022271), Dietzia sp. E1 (ACN62569), Dietzia sp. DQ12-45-1b (AEM66514), Prauserella rugosa NRRL B-2295 (CAB51024), Nocardioides sp. CF8 (AAK31348), and the fragment sequences of Dietzia sp. K44, Dietzia sp. ITRH 56 and Dietzia sp. H0B, showed great similarity between the sequences of P4, H0B and E1 (about 95%), and lower levels with K44 and ITRH56, 63% and 44% respectively. In addition, the multiple alignments between the deduced amino acid sequences identified three His boxes (Figure 2a-c) and an additional HYG-motif (Figure 2d). Both are commonly found among the alkane hydroxylases and have essential roles in its activity. Next to this, we found two CXCG Rub sequence motifs (Figure 2e, f), indicating an AlkG-type rubredoxin proteins (van Beilen et al., 2002, 2003).
In order to analyze the phylogenetic relationship between the deduced amino acid sequence of putative AlkB of P4 strain with other AlkB sequences of related actinobacteria, we compared different classes of alkane monooxygenase hydroxylase sequences available in NCBI data-base. The phylogenetic tree obtained (Figure 3) showed that the AlkB sequence of strain P4 is positioned in between other alkane monooxygenase sequences of Dietzia strains, separated from AlkB sequences of other actinomycetes, which is supported in 100% of the neighbour-joining trees generated. However, the evolutionary distance between the strain P4 AlkB sequence is apparently shorter with AlkB1 and AlkB2 than with AlkB3, AlkB4 and AlkB5. The robustness of the branching pattern tree was supported by 98% of maximum-parsimony and 100% of maximum-likelihood.
Growth and utilization of n-hexadecane as the sole carbon source
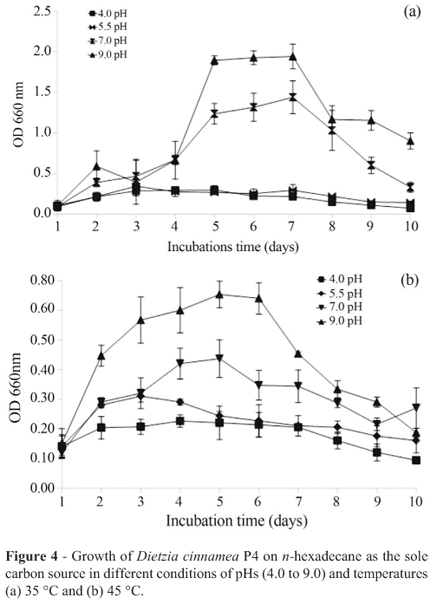
We evaluated the growth of strain P4 at different pHs (4.0, 5.0, 7.0 and 9.0), and temperatures (35 ºC and 45 ºC), using n-hexadecane as the sole carbon source (See Materials and methods). The time course of growth of strain P4 is shown in Figure 4. The strain was unable to grow in pH 4.0 and 5.0 at both temperatures; however it showed obvious growth at pH 7.0 and 9.0, at both temperatures. The strain showed initially slow growth, not presenting a significant increase up to 90 h of incubation, whereas after 90 h growth was exponential up to 120 h. Then, a stationary phase commenced, which lasted until 240 h, when cell viability started to decline. Similar growth kinetics was also described for growth in glucose as the sole carbon source at pH 7.0 and 35 ºC.
Expression of genes involved in the assimilation of n-hexadecane
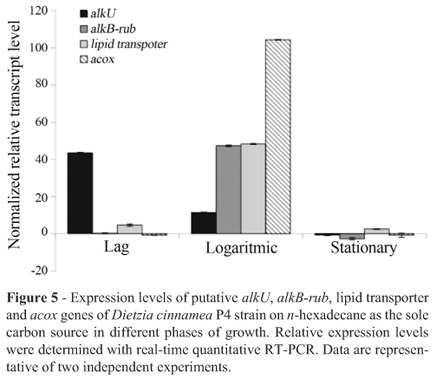
The expression of putative alkU, alkB-rub, the bifunctional ABC lipid transporter and acyl-CoA oxidase genes under the induction of n-hexadecane substrate during growth were analyzed by real-time RT-PCR. The level of transcripts of interest was compared to that obtained under induction by glucose as the sole carbon source, and the 16S rRNA gene was used for normalization for all conditions used (See Materials and methods). In the early stage of growth (the lag phase), the putative alkU gene showed the highest transcript level between the genes analyzed, being 43-fold that observed in the presence of glucose (Figure 5). In contrast, during the logarithmic growth phase, the level of the alkU regulator gene dropped to 11-fold, and to -0.5 in stationary phase.
The expression of alkB-rub and the bifunctional ABC lipid transporter genes in the presence of hexadecane showed increases of over 47-and 48-fold, respectively, during the late-exponential phase (Figure 5). During the early growth phase, while the alkane-1 monooxygenaserubredoxin showed a level close to zero, the bifunctional ABC lipid transporter showed 4.6-fold expression. A main destination for n-hexadecane in the cell is presumably its use in energy production, which uses the f-oxidation pathway (van Beilen and Witholt, 1994). In order to analyze whether there was any increase in activity of this catabolic pathway, we determined the levels of the acox (acyl-CoA oxidase) gene, which plays a key role in lipid metabolism. The expression levels of the acox gene during the logarithmic growth phase showed an increase of over 100-fold in the presence of n-hexadecane over that with glucose, whereas during the early and mid-stationary phases these expression levels remained low (below zero) (Figure 5).
Discussion
The soil bacterium D. cinnamea P4 has previously been isolated from microcosms containing oil-contaminated soil collected from an environmentally protected area of a tropical Atlantic forest (Biological Reserve of Poço das Antas -Brazil) (Evans et al., 2004). Von der Weid et al. (2007), in later experiments, showed the ability of strain P4 to grow in a wide range of n-alkanes of different sizes (C11 to > C36), oil-crude and aromatic hydrocarbons (e.g. benzene), revealing its potential to serve as an organism in bioremediation of petroleum hydrocarbons. Recently, the annotation of the genome of the bacterium D. cinnamea P4 was released by the NCBI site. This study was the first to describe a draft annotation of a Dietzia genome, which makes it highly relevant to advance the field of Dietzia biology. The genome revealed several interesting new genes and gene clusters with possible application in biotechnology and confirmed the presence of alk genes involved in degradation of n-alkanes (Procópio et al., 2012). In the present study, we showed the transcriptional profile of genes involved to use n-hexadecane compounds as the sole carbon and energy source under different conditions of pH (7.0-9.0) and temperature (35 ºC to 45 ºC).
Our bioinformatics analysis described a cluster of genes containing three putative ORFs directly involved in n-alkane transport and degradation, which is composed by one putative lipid transporter, one alkane monooxygenase and one TetR transcriptional regulator. The alkane monooxygenase described in this study is fused to a rubredoxin enzyme, which is necessary for the oxygenation reaction of n-alkane compounds. The unusual occurrence of an alkBrub natural fusion gene was also reported in Nocardioides sp. CF8 actinomycete (Hamamura et al., 2001) and Prauserella rugosa NRRL B-2295 (van Beilen et al., 2002). The presence of a monooxygenase as a fused polypeptide had already been described in Bacillus genus (Narhi and Fulco, 1987). The cytochrome P450 fatty acid monooxygenase from Bacillus megaterium was shown to consist of hydroxylase and reductase components on a single polypeptide (encoded by a single continuous gene), which can be cleaved by trypsin into the respective domains. Recently, the alkB-rub natural fusion gene was also reported in Dietzia sp. E1 (Bihari et al., 2011). The related Dietzia sp. E1 strain has a gene arrangement very similar to that described in D. cinnamea P4. In addition to the genes analyzed in this study, we identified other genes in strain P4 that showed homology and similar positions to that found in the chromosome of strain E1.
Although the degradation of differently-sized n-alkanes is reportedly ubiquitous in nature by different classes of alkane monooxygenases (AlkBs), the mechanisms have not been completely elucidated. A relationship between the AlkB protein structure and its preference for specific n-alkanes has been proposed. In the gram-negative Pseudomonas putida Gpo1, the hexagonal structure of AlkB builds a deep hydrophobic pocket, in which four conserved histidine residues allow a selective control over the specificity of the reaction catalyzed by the enzyme (van Beilen et al., 2005). In Gram-positive bacteria, like Rhodococcus genus, the presence of multiple alkane hydroxylases is a common feature (Whyte et al., 2002). This feature can be illustrated by the annotation of several alkB genes in a single gene cluster, as described in Rhodococcus ruber SP2B (Amouric et al., 2010). Despite the ability to degrade different size of n-alkanes, our genome annotation of D. cinnamea P4 identified only one putative alkane monooxygenase (Procópio et al., 2012). Phylogenetic analyses positioned the AlkB of strain P4 among those of Dietzia species, including the AlkB1 and AlkB2 alkane monoxygenase classes. The close relationship between the strain P4 and E1 AlkB sequences might indicate a similar role in hydrocarbon assimilation in respect of preferred sizes of alkanes (Bihari et al., 2011). The preference for medium-and long-chain hydrocarbons (> C8 and < C25) by D. cinnamea P4 was previously described by von der Weid et al. (2007).
The analysis of gene expression by real-time RT-PCR provided an excellent tool to determine the level of mRNA in response to environmental signals. Commonly, studies of gene expression of alk genes show an instantaneous transcriptional profile, taking into account the levels of transcripts involved in the assimilation of hydrocarbons after (short or long) exposure to the compounds (e.g. in late-exponential growth phase). Here, were evaluated the transcript levels of the putative strain P4 alk genes during the complete life cycle of the cells of D. cinnamea P4. We raised the question as to which levels of alk gene expression could be found during the early-phase of growth. In gramnegative bacteria, the regulator identified as the transcriptional regulator of the alk operon is alkS, belonging to the LuxR family, while in Actinobacteria, the transcriptional regulator identified by the alk genes expression is the TetR-type gene (alkU). Both regulators are normally found immediately downstream of the alkB gene (Whyte et al., 2002). In the annotated genome of D. cinnamea P4, the alk cluster genes were found in an arrangement where the puta-tive ulkU gene is immediately downstream of the alkB gene (Procópio et al., 2012). In this study, the expression of the alkU gene was analyzed by real-time RT-PCR during the complete growth cycle of P4, in the presence of n-hexadecane as the sole carbon source, and compared with the same conditions of growth, but using glucose as only carbon source. The obvious increase in expression levels of the putative alkU gene, after two hours in presence of n-hexadecane, and the subsequent increase of other alk genes in the logarithmic growth phase, indicated a clear role in inducing other alk genes, which were later expressed. The regulation of alk genes by the transcriptional regulator TetR is poorly described in the literature, and its action has only been related in gram-negative bacteria by the LuxRtype family transcriptional regulator (Moreno et al., 2007, 2009). The most studied transcriptional regulator responsible for alk operon induction is the AlkS protein, described in P. putida Gpo1 (Whyte et al., 2002). AlkS induces the expression of alk genes acting on n-alkanes from C5 to C16 (Sameshima et al., 2008). In a study conducted with the E1 strain, the expression levels of the TetR transcriptional regulator were analyzed only in late-exponential phase growth, which detected a low expression level of this gene in the presence of n-hexadecane compounds (Bihari et al., 2011).
The results of real-time RT-PCR in our study indicated the clear induction of the putative alkB-rub gene by n-hexadecane compounds (Figure 5). The increase during the late-exponential phase suggested transcription activated by the TetR-type transcriptional regulator. The hydroxylation reaction performed by alkane monooxygenase-rubredoxin is the key step in the assimilation of n-alkanes. Several studies on the induction of alkB genes by n-alkane compounds have taken into account the sizes of the preferred hydrocarbon chain class of alkane monoxygenases. Analysis of alkB-rub expression gene in Dietzia sp. E1 with different chain size n-alkanes showed the preference for C > 16 hydrocarbons (Bihari et al., 2011), while in Gordonia sp. SoCg had a higher level of alkB gene expression when grown in n-triacontane than n-hexadecane (Lo Piccolo et al., 2011). In addition, although our results did not indicate the possibility of activation of alk genes in the presence of hydrocarbons of different sizes, like the D. cinnamea P4 strain, the Dietzia sp. DQ12-45-1b as well P4 (although they have capabilities to use a wide range of n-alkanes) also preferentially degraded medium-and longchain hydrocarbons (Wang et al., 2011).
Although a probable transmembrane transporter for n-alkane compounds remains to be fully elucidated, studies with Mycobacterium album, Rhodococcus erythropolis S+14He and Acinetobacter sp., showed that, in the presence of n-hexadecane, there was an accumulation of intracellular inclusion bodies (Kennedy et al., 1975a,b). In addition, from thermodynamics, the presence of intracellular n-alkane compounds can only occur by an effective energy-dependent transport system (Kennedy et al., 1975b). In our experiments of real-time RT-PCR, the puta-tive lipid transporter gene of strain P4 showed an increase in expression levels during the late-exponential phase, probably under induction by transcriptional regulator AlkU. However, the expression of the lipid transporter genes during early and stationary phases showed elevated baseline levels when compared with other genes analyzed in this study. This was probably due to the need of such lipid carriers, at basal levels, required for the acquisition of n-alkanes compounds in a first contact. The presence of a lipid transporter along with the alkB gene is related by Liu et al. (2010) in Geobacillus sp. MH-1 strain. In addition, the location of a lipid transporter along of other genes involved in the assimilation of n-alkane compounds described in Dietzia sp. E1, with its concomitant induction by n-alkanes compounds, suggest its role in uptake of this compounds of environment (Bihari et al., 2011).
In conclusion, our results show that the P4 strain is able to grow using compounds of n-hexadecane as sole carbon source. The analysis of gene expression during the complete cycle growth suggests the role of the putative alkU TetR-family transcriptional regulator in inducing of the alkB-rub and lipid transporter genes during the lateexponential phase. This new information helps to elucidate the physiological behavior of the alk pathway, responsible for the degradation of oil-derived, which will allow a better use of this strain in bioremediation and biotransformation technologies.
Acknowledgments
The study was supported by PETROBRAS. Thanks to CNPq for providing a fellowship to L. Procópio to come over to the Groningen lab.
Submitted: April 1, 2012
Approved: July 23, 2012
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403-410.
- Amouric A, Quéméneur M, Grossi V, Liebgott PP, Auria R, Casalot L (2010) Identification of different alkane hydroxylase systems in Rhodococcus ruber strain SP2B, an hexane-degrading actinomycete. J Appl Microbiol 108:1903-1916.
- Atlas RM and Atlas MC (1991) Biodegradation of oil and bioremediation of oil spills. Curr Opin Biotechnol 2:440-443.
- Beal R and Betts WB (2000) Role of rhamnolipid biosurfactants in the uptake and mineralization of hexadecane in Pseudomonas aeruginosa J Appl Microbiol 89:158-168.
- Bihari Z, Szvetnik A, Szabó Z, Blastyák A, Zombori Z, Balázs M, Kiss I (2011) Functional analysis of long-chain n-alkane degradation by Dietzia spp. FEMS Microbiol Lett 316:100-107.
- Borsodi AK, Micsinai A, Rusznyák A, Vladár P, Kovács G, Tóth EM, Márialigeti K (2005) Diversity of alkaliphilic and alkalitolerant bacteria cultivated from decomposing reed rhizomes in a Hungarian soda lake. Microb Ecol 50:9-18.
- Borzenkov IA, Milekhina EI, Gotoeva MT, Rozanova EP, Beliaev SS (2006) The properties of hydrocarbon-oxidizing bacteria isolated from the oilfields of Tatarstan, Western Siberia, and Vietnam. Mikrobiologiia 75:82-89.
- Colquhoun JA, Mexson J, Goodfellow M, Ward AC, Horikoshi K, Bull AT (1998) Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie van Leeuwenhoek 74:27-40.
- Duckworth AW, Grant S, Grant WD, Jones BE, Meijer D (1998) Dietzia natronolimnaios sp. nov., a new member of the genus Dietzia isolated from an east African soda lake. Extremophiles 2:359-266.
- Evans FF, Rosado AS, Sebastián GV, Casella R, Machado PL, Holmström C, Kjelleberg S, Elsas JD, Seldin L (2004) Impact of oil contamination and biostimulation on the diversity of indigenous bacterial communities in soil microcosms. FEMS Microbiol Ecol 49:295-305.
- Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L (2007) Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci USA 104:5602-5607.
- Hamamura N, Yeager CM, Arp DJ (2001) Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl Environ Microbiol 67:4992-4998.
- Kennedy RS, Finnerty WR (1975a) Microbial assimilation of hydrocarbons. I. The fine-structure of a hydrocarbon oxidizing Acinetobacter sp. Arch Microbiol 102:75-83.
- Kennedy RS, Finnerty WR (1975b) Microbial assimilation of hydrocarbons. II. Intracytoplasmic membrane induction in Acinetobacter sp. Arch Microbiol 102:85-90.
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305:567-580.
- Liu YC, Li LZ, Wu Y, Tian W, Zhang LP, Xu L, Shen QR, Shen B (2010) Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour Technol 101:310-316.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25:402-408.
- Lo Piccolo L, de Pasquale C, Fodale R, Puglia AM, Quatrini P (2011) Involvement of an alkane hydroxylase system of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Appl Environ Microbiol 77:1204-1213.
- Moreno R, Marzi S, Romby P, Rojo F (2009) The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res 37:7678-7690.
- Moreno R, Ruiz-Manzano A, Yuste L, Rojo F (2007) The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol Microbiol 64:665-675.
- Narhi LO, Fulco AJ (1987) Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium J Biol Chem 262:6683-6690.
- Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 78:5636-43.
- Prince RC (2005) The microbiology of marine oil spill bioremediation. In: Ollivier B, Magot M (eds) Petroleum Microbiology. ASM Press, Washington DC, pp 317-336.
- Procópio L, Alvarez VM, Jurelevicius DA, Hansen L, Sørensen SJ, Cardoso JS, Pádula M, Leitão AC, Seldin L, van Elsas JD (2012) Insight from the draft genome of Dietzia cinnamea P4 reveals mechanisms of survival in complex tropical soil habitats and biotechnology potential. Antonie van Leeuwenhoek 101:289-302.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: Sequence visualization and annotation. Bioinformatics 16:944-945.
- Sabirova JS, Becker A, Lünsdorf H, Nicaud JM, Timmis KN, Golyshin PN (2011) Transcriptional profiling of the marine oil-degrading bacterium Alcanivorax borkumensis during growth on n-alkanes. FEMS Microbiol Lett 319:160-168.
- Sakai Y, Maeng JH, Tani Y, Kato N (1994) Use of long-chain n-alkanes (C13-C44) by an isolate, Acinetobacter sp. M-1. Biosci Biotechnol Biochem 58:2128-2130.
- Sameshima Y, Honda K, Kato J, Omasa T, Ohtake H (2008) Expression of Rhodococcus opacus alkB genes in anhydrous organic solvents. J Biosci Bioeng 106:199-203.
- Smits TH, Röthlisberger M, Witholt B, van Beilen JB (1999) Molecular screening for alkane hydroxylase genes in Gramnegative and Gram-positive strains. Environ Microbiol 1:307-317.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596-9.
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics, Chapter 2, Unit 2.3.
- Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB (2007) Identification of novel genes involved in longchain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl Environ Microbiol 73:3327-3332.
- van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13-21.
- van Beilen JB, Panke S, Lucchini S, Franchini AG, Röthlisberger M, Witholt B (2001) Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: Evolution and regulation of the alk genes. Microbiology 147:1621-1630.
- van Beilen JB, Neuenschwander M, Smits TH, Roth C, Balada SB, Witholt B (2002) Rubredoxins involved in alkane oxidation. J Bacteriol 184:1722-1732.
- van Beilen JB, Duetz WA, Schmid A, Witholt B (2003) Practical issues in the application of oxygenases. Trends Biotechnol 21:170-177.
- van Beilen JB, Smits TH, Roos FF, Brunner T, Balada SB, Röthlisberger M, Witholt B (2005) Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J Bacteriol 187:85-91.
- van Beilen JB, Witholt B (1994) Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174.
- Van Hamme JD, Singh A, Ward OP (2003) Recent advances in petroleum microbiology. Microbiol Mol Biol Rev 67:503-459.
- Van Hamme JD, Ward OP (2001) Volatile hydrocarbon biodegradation by a mixed-bacterial culture during growth on crude oil. J Ind Microbiol Biotechnol 26:356-362.
- von der Weid I, Marques JM, Cunha CD, Lippi RK, Dos Santos SC, Rosado AS, Lins U, Seldin L (2007) Identification and biodegradation potential of a novel strain of Dietzia cinnamea isolated from a petroleum-contaminated tropical soil. Syst Appl Microbiol 30:331-339.
- Wang XB, Chi CQ, Nie Y, Tang YQ, Tan Y, Wu G, Wu L (2011) Degradation of petroleum hydrocarbons (C6-C40) and cru-de oil by a novel Dietzia strain. Bioresour Technol 102:7755-7761.
- Whyte LG, Smits TH, Labbé D, Witholt B, Greer CW, van Beilen JB (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933-5942.
- Yumoto I, Nakamura A, Iwata H, Kojima K, Kusumoto K, Nodasaka Y, Matsuyama H (2002) Dietzia psychralcaliphila sp. nov., a novel, facultatively psychrophilic alkaliphile that grows on hydrocarbons. Int J Syst Evol Microbiol 52:85-90.
- Zhang Z, Hou Z, Yang C, Ma C, Tao F, Xul P (2011) Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour Technol 102:4111-4116.
Send correspondence to:
Publication Dates
-
Publication in this collection
09 Oct 2013 -
Date of issue
2013
History
-
Received
01 Apr 2012 -
Accepted
23 July 2012