Abstract
Microbial lipids, which are also known as single cell oils (SCO), are produced by oleaginous microorganisms including oleaginous bacteria, yeast, fungus and algae through converting carbohydrates into lipids under certain conditions. Due to its unique environment having extremely low temperature and anoxia, the Tibetan Plateau is amongst the regions with numerous rare ecotypes such as arid desert, salt marsh, alpine permafrost, hot spring, and lawn. By using a rapid, convenient screening method, we identified 31 strains of oleaginous microorganisms from different habitats in the Tibetan Plateau, which include wetlands, lawn, hot spring, alpine permafrost, and saline-alkali soil. Molecular identity analysis showed that they belong to 15 different species, 7 of which are reported for the first time as lipid-producing microorganisms, that is, Cladosporium sp., Gibberella fujikuro, Ochrobactrum sp., Plectosphaerella sp., Tilletiopsis albescens, Backusella ctenidia, and Davidiella tassiana. The distribution of the oleaginous microorganisms varies with habitats. 11 strains were found in hot spring (35.5%), 10 in farmland (32.3%), 6 in lawn (19.4%), 2 in sand (6.4%), 1 in wetland (3.2%), and 1 in permafrost (3.2%). Carbon utilization analysis indicated that most of these filamentous fungi can use xylose and carboxymethyl cellulose (CMC) as carbon source, where Backusella ctenidia, Fusarium sp. and Gibberella fujikuroi have the strongest capability.
oleaginous microorganisms; screening; the Tibetan Plateau; biodiversity
ENVIRONMENTAL MICROBIOLOGY
Biodiversity of the oleaginous microorganisms in Tibetan Plateau
Shi Lin Li; Qiang Lin; Xin Ran Li; Hui Xu; Yun Xi Yang; Dai Rong Qiao; Yi Cao* * Corresponding Author. Mailing address: College of Life Sciences, Sichuan University, Sichuan, 610064, P. R. China..; Tel: 86 28 85412842 Fax: 86 28 85412842.; E-mail: geneium@scu.edu.cn
Microbiology and Metabolic Engineering Key Laboratory of Sichuan Province, College of Life Science, Sichuan University, Chengdu, 610064, P.R. China
ABSTRACT
Microbial lipids, which are also known as single cell oils (SCO), are produced by oleaginous microorganisms including oleaginous bacteria, yeast, fungus and algae through converting carbohydrates into lipids under certain conditions. Due to its unique environment having extremely low temperature and anoxia, the Tibetan Plateau is amongst the regions with numerous rare ecotypes such as arid desert, salt marsh, alpine permafrost, hot spring, and lawn. By using a rapid, convenient screening method, we identified 31 strains of oleaginous microorganisms from different habitats in the Tibetan Plateau, which include wetlands, lawn, hot spring, alpine permafrost, and saline-alkali soil. Molecular identity analysis showed that they belong to 15 different species, 7 of which are reported for the first time as lipid-producing microorganisms, that is, Cladosporium sp., Gibberella fujikuro, Ochrobactrum sp., Plectosphaerella sp., Tilletiopsis albescens, Backusella ctenidia, and Davidiella tassiana. The distribution of the oleaginous microorganisms varies with habitats. 11 strains were found in hot spring (35.5%), 10 in farmland (32.3%), 6 in lawn (19.4%), 2 in sand (6.4%), 1 in wetland (3.2%), and 1 in permafrost (3.2%). Carbon utilization analysis indicated that most of these filamentous fungi can use xylose and carboxymethyl cellulose (CMC) as carbon source, where Backusella ctenidia, Fusarium sp. and Gibberella fujikuroi have the strongest capability.
Key words: oleaginous microorganisms, screening, the Tibetan Plateau, biodiversity
INTRODUCTION
Lipid is the transient and storage form of energy needed for metabolism. However, it is not only the energy provider for an organism but also the important building block. For example, phospholipid is one of the most important compounds of biomembrane. Under certain conditions, some microorganisms transform carbohydrate, hydrocarbon and normal lipid into lipids within the cells (15). Previous studies suggested that oleaginous microorganisms are mainly bacteria, yeast, filamentous fungi and microalgae. It was also reported that the lipid content in microalgae, yeasts and filamentous fungi was higher (70%-90%) than that in bacteria (20%-50%) (19).
The Tibetan Plateau is referred to as "the third pole" of the Earth due to its unique natural and geographical characteristics. Extremely harsh conditions, such as low temperature, oligotrophy, hypoxia, and strong ultraviolet and magnetic radiation, breed abundant extreme microorganisms. These extreme microorganisms have special genetic characteristics of physiological and biochemical adaptation mechanisms that enable them to survive in such a bad environment. Up till now, there has been no report on the biodiversity of the oleaginous microorganisms in the Tibetan Plateau.
Previous studies indicated that most of the oleaginous microorganisms can only utilize glucose as carbon source to produce lipids. However, the cost of the biodiesel produced by these microorganisms is so high that its sustainable development is limited. As a result, one of the most important questions in the microbial fermentation of lipids is substrate utilization. Chen et al. identified that some oleaginous yeasts could tolerate the hydrolyzates of lignocellulose while producing microbial lipids (3, 5). However, whether this is true for filamentous fungi remains elusive.
The conventional method used for lipid determination is Sudan Black B or Sudan III staining (18), however, the proportion of false-positive is relatively high, and the amount of lipid particles is not consistent with lipid content (7). At present, many studies on oleaginous microorganisms focus on the screening of unknown strains, the discovery of new oleaginous microorganisms and the optimization of fermentation conditions (3, 8, 9, 12, 14). However, there are very few reports on the biodiversity of the oleaginous microorganisms in different habitats (7, 13). In this study, we established a new method based on different growth rates in medium lack of carbon and identified some new oleaginous microorganisms from different habitats in the Tibetan Plateau. The study on the biodiversity of the oleaginous microorganisms enriched and provided the distribution of oleaginous microorganisms in different habitats in the Tibetan Plateau.
MATERIALS AND METHODS
Soil sample collection
26 soil samples were collected 5-20 cm below the surface from hot spring, permafrost, wetland, sand, lawn, saline-alkali soil, high-radiation soil and farmland in Haibei, the Tibetan Plateau, and were stored at 4°C until use.
Reagents
Ex-Taq polymerase and PMD18-T Plasmid were obtained from TaKaRa (Japan). All other reference substances and chemicals were purchased from Sinopharm Chemical Reagent (China), and were of analytical grade unless otherwise specified.
Enrichment of oleaginous microorganisms: 1g of soil sample was added into a 250 mL flask containing 50 mL sterilized enrichment medium to obtain a mixture. The enrichment medium contains (g/L) glucose 100, yeast extract 1, NH4Cl 1, KH2PO4 2, MgSO4.7H2O 0.75, CaCl2.2H2O 0.05, ZnSO4.7H2O 0.01, FeCl3.6H2O 0.01 and Na2HPO4 1, and it has a pH of 7.4. The mixture was cultured at 28°C, 180rpm for 48h to allow the amount of oleaginous microorganisms and the content of lipids to reach a certain levels.
Screening and isolation of oleaginous microorganisms
1 mL enriched sample was serially (1:10) diluted with sterilized water, and then 0.1 mL of the diluent was spread evenly on a screening plate. The screening medium does not have carbon, but contains (g/L): yeast extract 1, NH4Cl 1, KH2PO4 2, MgSO4.7H2O 0.75, CaCl2.2H2O 0.05, ZnSO4.7H2O 0.01, FeCl3.6H2O 0.01 and Na2HPO4 1. The screening medium was adjusted to a pH of 7.4 and was kept in an incubator at 28°C for 1-4 days. The strains that appeared the earliest and grew the fastest were picked for further study.
Activation and fermentation of oleaginous microorganisms
Of the oleaginous microorganisms obtained, filamentous fungi were activated on PDA for 72 h, yeasts on YEPD for 48h and bacteria on LB for 48 h before being added into a 250 mL flask containing 100 mL fermentation medium (glucose 60, yeast extract 1, NH4Cl 1, KH2PO4 2, MgSO4.7H2O 0.75, CaCl2.2H2O 0.05, ZnSO4.7H2O 0.01, FeCl3.6H2O 0.01, Na2HPO4 1g/L, pH 7.4.) The medium mixed with the oleaginous microorganisms was cultured at 28°C, 200 rpm for 6 days, where triplicate samples were set up to determine biomass, dry weight and lipid content.
Determination of biomass (dry weight) of oleaginous microorganisms
Zymotic fluid was spun down at 6,000 g for 10 minutes to collect thalli before it was washed twice with sterilized water and centrifuged again. The thalli were kept at 80°C for 24 h before biomass (dry weight) was determined.
Extraction of lipid compounds
Lipids were extracted by the advanced Bligh and Dyer method (1). Briefly, 100 mL of the zymotic fluid was centrifuged at 5,000 g for 10 min to obtain thalli. The collected thalli were then transfered to a 50 mL centrifugal tube and washed twice with sterilized water. 15 mL of 4 M HCl were added to the thalli and the mixture was kept at room temperature for 30 min before it was dipped in liquid nitrogen for 10 min and subsequently in boiling water for 10 min. This freezing/thawing process was repeated 3 times in order to break up the cells. 30 mL chloroform/methanol (1:1) was added into the tube, shaked vigorously with a vortex oscillator and then centrifuged at 5,000 g for 10 min. The lipid-containing chloroform layer (the lower layer) was dried in a decompression device before it was weighed to obtain the content of lipids.
Iodine value determination of microbial lipids
The advanced Hanus method was used (7).
Saponification value determination of microbial lipids
Lipids were mixed with excess amount of potassium hydroxide ethanol solution for saponification. With phenolphthalein as an indicator, hydrochloric acid standard solution was used to titrate the remaining potassium hydroxide. Blank control was performed at the same time. The amount of potassium hydroxide consumed in saponification marked the saponification value of the microorganism lipids.
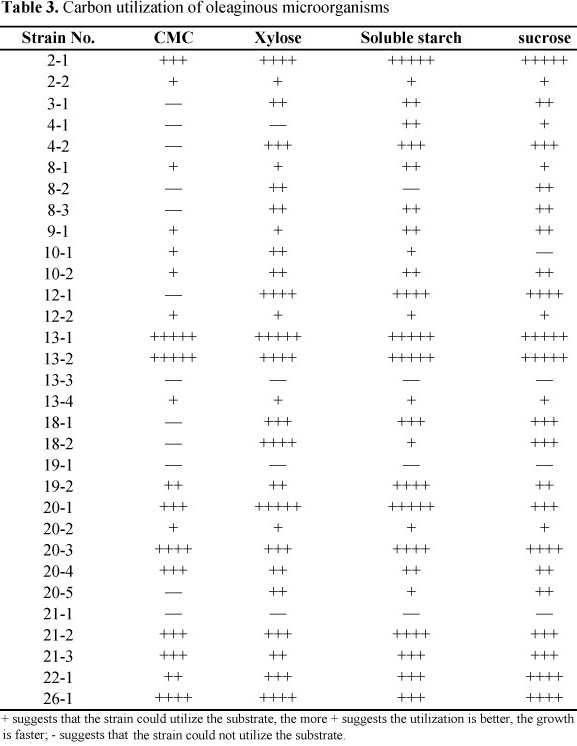
Analysis of carbon utilization of oleaginous microorganisms
The substrate-using medium formula was as follows (g/L): NH4Cl 5 g, KH2PO4 2 g, MgSO4.7H2O 0.75 g, CaCl2.2H2O 0.05 g, ZnSO4.7H2O 0.01 g, FeCl3.6H2O 0.01 g, Na2HPO4 1 g, agar 15 g, pH 7.4. Besides, CMC, xylose, soluble starch and sucrose as the single carbon source respectively were added and the final concentration was 10 g/L. The isolated strains were inoculated into these media. The result with "+" suggests that a strain could utilize the substrate. The more "+" a result is labeled with, the better the utilization is, and the faster the growth is. The symbol "-" suggests that a strain could not utilize the substrate.
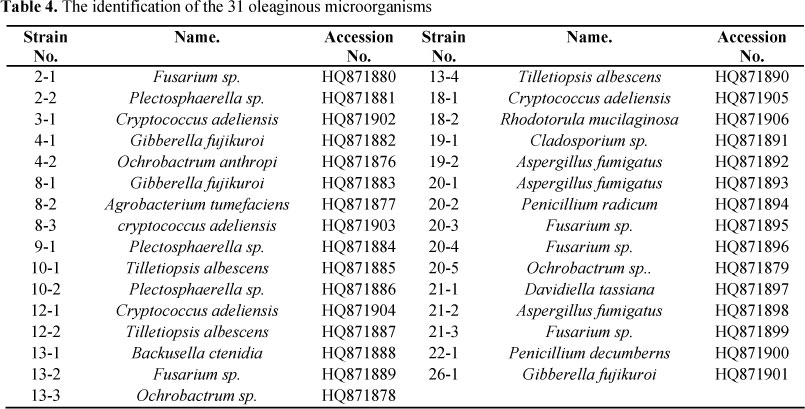
Molecular identification of oleaginous microorganisms
The genomic DNA of each strain was extracted by the SDS-Proteinase K-CTAB method (16). All DNA samples were treated with RNase A and examined on ethidium bromide-stained 1% agarose gels. Primers NL1 and NL4 were used to amplify 26S D1/D2 fragment of yeast (6); primers EF3 and EF4 to amplify 18S sequence of fungi (17); and primers F27 and R1492 to amplify 16S sequence of bacteria (10). The amplified PCR products were sequenced by Invitrogen Corporation (Shanghai, China) and the results were blasted in NCBI. For PCR primers, see Table 1.
Statistical analysis
All the experiments were performed in triplicates and the data were analyzed using one way analysis of variance (ANOVA). Differences with p<0.05 were considered statistically significant.
RESULTS
Isolation of oleaginous microorganisms
31 strains were identified at the first screening, based on the time they appeared and their growth rate on a screening plate without carbon (Table 2). A series of parameter determination was performed, including biomass, lipid yield, lipid content, iodine value and saponification value determination (Table 2). All the data obtained were expressed as the average of three determinations. As shown in Table 2, the initial lipid content of the microorganisms was 15%-45%, which suggests the reliability of the isolation method.
Carbon utilization of oleaginous microorganisms
As shown in Table 3, nearly all filamentous fungi can utilize xylose, CMC, starch and sucrose as carbon source; while yeasts and bacteria can only utilize xylose, starch and sucrose but not CMC. Strains 13-1, 13-2, 20-3 and 26-1 can utilize CMC well, suggesting the potential of using agricultural waste in microbial lipid production.
Molecular identification of oleaginous microorganisms
The results of 16S, 18S and 26S sequencing and BLAST indicated that 31 strains we identified belong to 15 different species (Table 4). Seven out of 15 are reported for the first time as lipid-producing microorganisms, that is, Cladosporium sp., Gibberella fujikuro, Ochrobactrum sp., Plectosphaerella sp.,Tilletiopsis albescens, Backusella ctenidia, and Davidiella tassiana.
DISCUSSION
Isolation of oleaginous microorganisms
The basic mechanism of lipid accumulation in microorganisms has been well studied. When the culture medium contains sugar but low nitrogen, lipid accumulates (2, 4, 8, 11). In order to isolate oleaginous microorganisms, Sudan Black B staining is usually used to determine lipid content (18). However, this method only roughly indicates the presence of microbial lipids, and it provides no quantification of the lipid content. So there are many false-positive results (3, 7) that make isolation difficult. In the present study, oleaginous microorganisms were firstly enriched in high C/N medium, allowing them to accumulate a certain amount of lipids. Then strains were selected based on the time they appeared and their growth rate on medium without carbon. Results (Table 2) showed that the initial lipid content was 15%-45%. The time the stain appears is coherent with the lipid content. The earlier it appears, the more lipids it contains. This indicated the reliability of this method. Moreover, the results of iodine value and saponification value determination (Table 2) suggest that the compounds of microbial lipids are similar to those of vegetable oil.
Biodiversity of the oleaginous microorganisms
Previous studies showed that many species of microorganisms can accumulate lipids efficiently, such as Schizochytrium sp., Arthrobacter sp., Bacillus sp., Candida sp., Cryptococcus sp., Aspergillus sp., etc. (15) Pan et al. obtained 13 different oleaginous yeasts from soil (7), although most of them had already been reported. Oleaginous microorganisms we obtained belong to 15 different species (Table 4), among which 8 species have been reported and the other 7 species are newly discovered, that is, Cladosporium sp., Gibberella fujikuro, Ochrobactrum sp., Plectosphaerella sp., Tilletiopsis albescens, Backusella ctenidia, and Davidiella tassiana. Our data indicated that the biodiversity of oleaginous microorganisms in special habitats in the Tibetan Plateau is extremely rich.
Relationship between oleaginous microorganisms distribution and habitats
The environment with high C/N ratio is good for lipid accumulation. 31 oleaginous microorganisms we obtained are from 6 different habitats (Table 2). As shown in Table 2, 11 strains (35.5%) were isolated from hot spring samples, most of which are filamentous fungi; 10 strains (32.3%) were isolated from farmland samples, some of which are yeasts and bacteria; 6 strains (19.4%) were isolated from lawn samples. However, there are a few oleaginous microorganisms in wetland, sand and permafrost samples, while there are no oleaginous microorganisms in alkali soil and highly radiated soil. It suggests that oleaginous microorganisms may distribute in a nutrient-rich environment and high temperature has a positive effect on the lipid accumulation of oleaginous microorganisms.
Biodiversity of the oleaginous microorganisms on carbon utilization
Most of the oleaginous microorganisms reported utilize glucose to produce lipids (8, 9, 14). However, using glucose in microbial lipid production will increase the cost greatly, which limits its application. Previous studies reported that some oleaginous yeasts could use the hydrolyzates of methyl cellulose in lipid production (3, 5). However, there are no relevant reports on filamentous fungi. The substrate utilization of 31 strains we obtained indicates that most of the filamentous fungi can utilize xylose and CMC, among which Backusella ctenidia, Fusarium sp., Gibberella fujikuroi can utilize xylose and CMC well, suggesting the potential of using agricultural waste in microbial lipid production. The results in this current study laid a solid foundation for using these strains in producing microbial lipid from agricultural wastes.
ACKNOWLEDGEMENTS
This work was supported by the National Special Basic Research of China (No. SB2007FY400) and the National Basic Research Program of China (No. 2009CB125910).
Submitted: March 18, 2011
Returned to authors for corrections: April 21, 2011
Approved: January 16, 2012
- 1. Bligh, E.G.; Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Physiol. Pharmacol. 37(8), 911-917.
- 2. Botham, P.A.; Ratledge, C. (1979). A biochemical explanation for lipid accumulation in Candida 107 and other oleaginous micro-organisms. J. Gen. Microbiol. 114(2), 361-375.
- 3. Chen, X.; Li, Z.H.; Zhang, X.X.; Hu, F.X.; Dewey, D.Y.; Bao, J. (2009). Screening of Oleaginous Yeast Strains Tolerant to Lignocellulose Degradation Compounds. Appl. Biochem. Biotechnol. 159(3), 591-604.
- 4. Evans, C.T.; Scragg, A.H.; Ratledge, C. (1981). Regulation of citrate efflux from mitochondria of oleaginous and non-oleaginous yeasts by adenine nucleotides. Eur. J. Biochem. 132(3), 609-615.
- 5. Hu, C.M,.; Zhao, X.; Zhao, J.; Wu, S.G.; Zhao, Z.B. (2009). Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresource. Technol 100(20), 4843-48471.
- 6. Kurtzman, C.P.; Robnett, C.J. (1998). Identification and phylogeny of ascomicetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequence. Anton. Leeuw. Int. J. G. 73(4), 331-371.
- 7. Li, X.P.; Deng, F.Y.; Li, S.; Wei, L.; Gui, G.C.; Zhi, Q.L. (2009). Isolation of the Oleaginous Yeasts from the Soil and Studies of Their Lipid-Producing Capacities. Food. Technol. Biotech. 47(2), 215-220.
- 8. Li, Y.H.; Zhao, Z.B.; Bai, F.W. (2007). High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme. Microb. Technol. 41(3), 312-317.
- 9. Loffhagen, N.; Hartig , C.; Harms, H. (2006). Impact of membrane fatty acid composition on the uncoupling sensitivity of the energy conservation of Comamonas testosterone ATCC 17454. Appl. Microbiol. Biot. 70(5), 618-624.
- 10. Macrae, A. (2000). The use of 16S rDNA methods in soil microbial ecology. Braz. J. Microbiol. 31(2), 77-82.
- 11. Palmieri, L.; Palmieri, F.; Runswick, M.J. (1996). Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. Febs. Lett. 399(3), 299-302.
- 12. Papanikolaou, S.; Komaitis, M.; Aggelis, G. (2004). Single cell oil(SCO) production by Mortierella isabellina grown on high-sugar content media. Bioresour. Technol. 95(3), 287-291.
- 13. Patnayak, S.; Sree, A. (2005). Screening of bacterial associates of marine sponges for single cell oil and PUFA. Lett. Appl. Microbiol. 40(5), 358-363.
- 14. Ratledge, C. (2002). Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. T. 30(6), 1047-1050.
- 15. Ratledge, C.; Wynn, J. (2002). The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 51, 1-44.
- 16. Sambrook, J.; Russel, D.W. (2001). Molecular cloning: A laboratory manual. 3rdedn. CSH Laboratory Press, Cold Spring Harbor, New York.
- 17. Smit, E.; Leeflang, P.; Glandorf, B.; Van Elsas, J.D.; Wernars, K. (1999). Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microb. 65(6), 2614-2621.
- 18. Thakur, M.S.; Prapulla, S.G.; Karanth, N.G. (1989). Estimation of intracellular lipids by the measurement of absorbance of yeast cells stained with Sudan Black B. Enzyme. Microb. Tech. 11(4), 252-254.
- 19. Xin, M.; Yang, J.M.; Xu, X.; Zhang, L.; Nie, Q.J.; Xian, M. (2009). Biodiesel production from oleaginous microorganisms. Renew. Eners. 34(1), 1-5.
Publication Dates
-
Publication in this collection
07 Aug 2012 -
Date of issue
June 2012
History
-
Received
18 Mar 2011 -
Accepted
16 Jan 2012 -
Reviewed
21 Apr 2011