Abstract
Food handlers, an important factor in food quality, may contain bacteria that are able to cause foodborne disease. The present study aimed to research coagulase-negative (CNS) and -positive staphylococci (CPS) in 82 food handlers, analyzing nasal and hand swabs, with identification of 62 CNS (75.6%) and 20 CPS strains (24.4%). Staphylococcal enterotoxins genes were investigated by PCR. In 20 CPS strains, 19 were positive for one or more genes. The percentage of CNS presenting genes for enterotoxins was high (46.8%). Despite of the staphylococcal species, the most common gene was sea (35.4%), followed by seh and sej (29.2%). The detection of new staphylococcal enterotoxins (SEs) genes showed a higher pathogenic potential in this genus. The presence of these gene points out the importance of CNS not only as contaminant bacteria but also as a pathogen.
Staphylococcal enterotoxins; coagulase-negative staphylococci; S. aureus; food handler
FOOD MICROBIOLOGY
Detection of enterotoxin genes of Staphylococcus sp isolated from nasal cavities and hands of food handlers
Rall, V.L.M.I,* * Corresponding Author. Mailing address: Department of Microbiology and Immunology, Institute of Biosciences, Sao Paulo State University -UNESP. Post Office Box 510. 18618-000, Botucatu, Sao Paulo, Brazil.; Tel. (+5514) 3811-6240 Fax (+5514) 3815-3744.; Email: vlmores@ibb.unesp.br ; Sforcin, J.M.I; Augustini, V.C.M.I; Watanabe, M.T.I; Fernandes Jr., A.I; Rall, R.II; Silva, M.G.III; Araújo Jr., J.PI
IDepartamento de Microbiologia e Imunologia, Instituto de Biociências, Universidade Estadual Paulista, Botucatu, SP, Brasil
IIFaculdade de Tecnologia, Botucatu, SP, Brasil
IIIDepartamento de Patologia, Universidade Estadual Paulista, Botucatu, SP, Brasil
ABSTRACT
Food handlers, an important factor in food quality, may contain bacteria that are able to cause foodborne disease. The present study aimed to research coagulase-negative (CNS) and -positive staphylococci (CPS) in 82 food handlers, analyzing nasal and hand swabs, with identification of 62 CNS (75.6%) and 20 CPS strains (24.4%). Staphylococcal enterotoxins genes were investigated by PCR. In 20 CPS strains, 19 were positive for one or more genes. The percentage of CNS presenting genes for enterotoxins was high (46.8%). Despite of the staphylococcal species, the most common gene was sea (35.4%), followed by seh and sej (29.2%). The detection of new staphylococcal enterotoxins (SEs) genes showed a higher pathogenic potential in this genus. The presence of these gene points out the importance of CNS not only as contaminant bacteria but also as a pathogen.
Key words: Staphylococcal enterotoxins, coagulase-negative staphylococci, S. aureus, food handler.
INTRODUCTION
Food handlers contribute to food safety, being potential sources of bacteria that causes foodborne diseases due to the introduction of pathogens during its processing, distribution and manipulation (1).
Staphylococcus aureus, produces enterotoxins and is considered one of the greatest causes of intoxication although its found persistently or temporarily in human nasal microbiota, without causing any symptoms. The presence of these bacteria in food occurs frequently due to inappropriate manipulation of food by carriers of this microorganism (7). The nasal mucosa has been described as the most important source of propagation, being colonized in the first days of life Among staphylococci, S. aureus is considered the most important to man and can be found in the nasal mucosa in 20 to 50% of adults in cutaneous pleats, armpits, as well as in inguinal and perineal areas (33).
The staphylococcal enterotoxins are considered superantigens, characterized by simultaneous connections to the major complex of histocompatibility class II in an antigen presenting cell and T cell receptors, without the presence of specific antigens, resulting in systemic effects such as high fever, vomiting, diarrhea, as well as hepatic and renal dysfunctions (6).
Classic antigenic SE have been identified as SEA, SEB, SEC1, SEC2, SEC3, SED and SEE (4). Ren et al. (26) have sequenced the gene of toxin H. In 1998, Munson et al. identified and characterized seg and sei genes and Zhang et al. (34) found the gene sej in the same plasmid that encoded sed. Recently, several other toxins have been described and their genes have been sequenced, known as SEK, SEL, SEM, SEN, SEO, SEP, SEQ, SER and SEU (8,11, 14, 21,22,23). Besides SEH, SEI and SEG, which present emetic activity (17,30), the involvement of other SEs in foodborne outbreaks is not clear yet. Other coagulase-positive staphylococci (CPS) species, such as S. hycus, S. intermedius and several coagulase-negative staphylococci (CNS), have also been involved in cases of outbreaks (32).
The present work aimed to investigate the presence of coagulase-positive and -negative staphylococci in nasal mucosa and hands of food handlers at industrial kitchens in the city of Botucatu and to detect the genes responsible for SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI and SEJ production.
MATERIAL AND METHODS
Material for analysis
Botucatu is a small town and there are only 3 industrial kitchens with a significant number of workers. Apparently, they are clean but didn't follow GHPs or HACCP implemented.
Samples were collected from the hands (interdigital region, indexfingers, thumbs and palms of both right and left hands) and anterior nares of 82 food handlers, distributed among 3 kitchens in the city, with a moist swab (saline), during meal preparation. One swab was used in each region.
The swabs were streaked on Baird-Parker plates (Difco) immediately after collection. As soon as possible (until 1 hour), these plates were incubated at 35ºC for 48 hours in our laboratory.
Staphylococcal isolation and identification (13)
After incubation, five black colonies from each plate (presence and absence of halo) were identified. The screening tests used were Gram staining and production of catalase and coagulase. The coagulase-positive strains were submitted to the kit "Staphytect Test Dry Spot" (Oxoid). The two positive clumping species were submitted to the VP test (S. aureus-positive and S intermedius-negative).
The coagulase-negative staphylococcal isolates were identified using API Staphy and according to Kloos and Bannerman (10), with ornithine decarboxylase presence, β hemolysis production, urea degradation, novobiocin resistance and anaerobic thioglycolate growth. Sensibility tests for bacitracin (0.04U) and furazolidone (100 µg) were performed before API, in order to separate CNS from Kocuria, according to Bannerman & Peacock (2).
DNA isolation
For DNA isolation, a GFX commercial kit (GE Healthcare) was used, according to supplier instructions.
Detection of genes encoding staphylococcal enterotoxins
Primers used in the detection of SE genes are listed in Table 1.
PCR amplifications were composed of 2.5 µL PCR Buffer 10x (Invitrogen), 1.0 µM of MgCl2 (Invitrogen), 200 µM dNTP (Invitrogen), 1 U of Taq DNA Polymerase (Invitrogen), 10 picomols of each primer, 3 µL of the DNA sample, and sterile ultrapure water in order to reach 25 µl (qsp) (Milli-Q Plus, Millipore).
PCR protocol was performed in PTC-100 (MJ Research, Inc., USES) using the following amplification cycles: initial denaturation for 5 minutes at 94ºC and 30 cycles at 94ºC for 2 minutes for denaturation and 72ºC for 1 minute for extension. The various temperatures used in the annealing step are shown in Table 1. Final extension was performed at 72ºC for 5 minutes. The PCR-amplified samples were analyzed by electrophoresis for 30 minutes at 125V (Electrophoresis Power Supply Model EPD 600 - Amersham-Pharmacia Biotech Inc.) using agarose gel (1.5% -Sigma Aldrich) added to ethidium bromide (0.5 mg/ml -Invitrogen) in TBE (0.09 M Tris-HCl, 0.09 M boric acid, 2 mM EDTA, pH 8.3). A 50 bp ladder was used (Ladder -Amersham -Pharmacia Biotech Inc.). The results were visualized by employing an image analyzer (Alphaimager -Alpha Esasy FC Software -AlphaInotech Corporation).
Positive controls used were S. aureus ATCC 13565 (SEA), ATCC 14458 (SEB), ATCC 19095 (SEC), FRI 361 (SED, SEG, SEI and SEJ), ATCC 27664 (SEE) and FRI 137 (SEH).
One sample of each SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI, and SEJ amplicon were sequenced and partial sequences were confirmed to correspond to the GenBank accessions M18970, M11118, X05815, M28521, M21319, AY920261, U11702, AY920268, and AB075606 respectively.
RESULTS AND DISCUSSION
Food handler may be a vector of foodborne disease spreading, due to inadequate personal hygiene or cross contamination (3).
Several authors have studied the presence of S. aureus in the nasal cavities of food handlers. Vanderbergh et al. (31) reported that the isolation of S. aureus from nasal cavities, could vary from 20 to 55% in a healthy adult population. These data are in agreement with the present work since the microorganism was isolated nasally in 15 (22.1%) food handlers out of a total of 68.
After the discovery of the new enterotoxins, the percentage of enterotoxigenic or potentially enterotoxigenic S. aureus increased because almost the totality (95%) isolated from nasal cavities and hands of food handlers was positive for the presence of genes of at least one enterotoxin; however, considering only the classic ones (SEA to SEE), the number falls to 13 (68.4%). Rosec and Gigaud (27) also observed an increased number of enterotoxigenic isolates with the discovery of the new toxins because 30% of the strains in their study had genes for the classic toxins compared with 57% for the new ones.
According to Table 2, 13 strains of S. aureus presented at least one gene for classic enterotoxin production and sea was the most observed, occurring in 9 isolates (69.2%), followed by 4 positive strains for see (30.8%), 3 for sec (23.1%) and 1 (7.7%) for seb, whereas SED-encoded genes were not observed. Nashev et al. (18) observed low positivity for sed gene, only 4.5% among the food handlers. Toxin A has been described as the most common by many other authors (19, 24).
The gene seg was observed in 13 (27.1%) of the 48 staphylococcal samples and in 84.6% of the cases, it was associated with the sei gene, which occurred in 11 strains (22.9%). Similar values were described by Rosec and Gigaud (27), and seg and sei were associated to 80.6% of the 155 positive isolates. Nashev et al. (18) studied 44 S. aureus strains and 12 (27.3%) were positive for the genes seg and sei, a value very similar to that found in the present work. These genes are frequently found together because they are located in sequential positions, in a DNA fragment of 3.2 Kb (8).
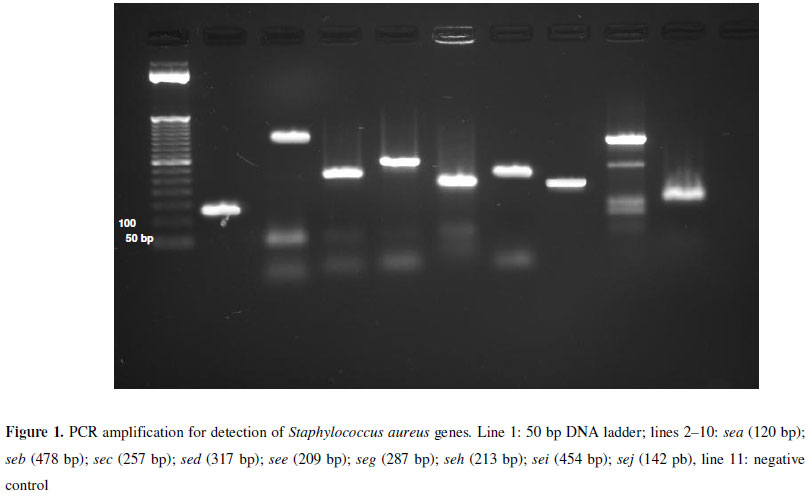
Among the CNS, 37 (45,1%) strains of S. warneri, 23 (28%) of S. epidermidis, 1 (1,2%) of S. capitis and 1 (1,2%) of S. xylosus were identified, in a total of 62 strains and out of 29 (46.8%) were positive for at least one gene encoded enterotoxin, the most common was sej (31%), followed by sea and seh (24.1%), seb (17.2%), see (13.8%), sec (6.9%) and, finally, seg and sei, with 3.4% each. Figure 1 shows the patterns according to the primers used in PCR reaction.
Several studies demonstrated the presence of different species of CNS-producing enterotoxins and/or others encoding these genes. The number of CNS enterotoxin-encoding genes in the present work can be considered high, 46.8% (29 positive). A much smaller prevalence was observed by Crass and Bergdoll (5), who found 16.5% of 2000 CNS strains. However, if only the classic enterotoxins are taken into account, exactly as investigated by Crass and Bergdoll (5), that number falls to 19.4% (12 positive strains for sea to see), a value close to that observed by these authors. In the present work, see was observed in S. warneri and S. capitis, whereas S. xylosus did not present genes for the production of any of the investigated toxins. However, the presence of those genes don't indicate the capacity of enterotoxin production in amounts enough to cause clinical signals (15).
In Brazil, 63,2% strains of S. epidermidis and 84,6% of S. cohnii isolated from food handlers involved in foodborne diseases were enterotoxin producers (28). The enterotoxin production by CNS was also studied by Rapini et al. (25) who observed S. epidermidis and S. cohnii isolated from food handlers, producing SEA - SED, in 96,4% from 28 pools of these species. According to Lamaita et al (12), 41,3% of CNS produced isolated from raw milk produced some SE, including S. epidermidis, S. sciuri, S. cohnii, S. hyicus, S. schleiferi, S. intermedius e S. delphini. Stamford et al. (29) observed the enterotoxin production by S. chromogenes (50%) and other 2 strains of CNS, but these species were not identified.
According to our results, S. aureus and CNS were isolated from food handlers, with a predominance of CNS. Genes that encode the production of classic toxins (SEA to SEE) and the most recent ones (SEG, SEH, SEI and SEJ) for CNS and CPS were also observed. The research on the new enterotoxins considerably increased the percentage of enterotoxigenic strains in both groups, increasing the pathogenic potential of this group of bacteria. CNS was always considered a food contaminant, without receiving attention. However, with the discovery of their capacity to produce enterotoxins, the role of those microorganisms should be reviewed, as well as and their importance should be taken into account. Besides, further investigations are needed to verify the production of these new enterotoxins and their real role in foodborne disease.
ACKNOWLEDGEMENTS
The authors thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for support and Dr. Ivano de Filippis (Fundação Oswaldo Cruz/Rio de Janeiro/RJ) for providing toxin-producing S. aureus reference strains.
Submitted: May 08, 2008; Returned to authors for corrections: April 20, 2009; Approved: September 28, 2009.
- 1. Angelillo, I.F.; Viggiani, N.M.A.; Rizzo, L.; Bianco, A. (2000). Food handlers and foodborne diseases: knowledge attitudes and reported behavior in Italy. J. Food Prot., 3, 381-385.
- 2. Bannerman, T.L.; Peacock, S.J. (2007). Staphylococcus, Micrococcus and other catalase-positive Cocci. In: Murray, P.R.; Baron, E.J.; Jorgensen, J.H.; Landry, M.L.; Pfaller, M.A. (eds). Manual of Clinical Microbiology, Ed. 9th, ASM Press, Washington, DC, p.390-411.
- 3. Bass, M.; Ersun, A.S.; Kivanç, G. (2006). The evaluation of food hygiene knowledge, and practices of food handlers in food businesses in Turkey. Food Control, 17, 317-322.
- 4. Bergdoll, M.S.; Robbins, R.N. (1973). Characterization of types staphylococcal enterotoxins. Journal of Milk Food Technology, 36, 610-2.
- 5. Crass, B.A.; Bergdoll, M.S. (1986). Involvement of coagulase negative staphylococci in Toxic Shock Syndrome. J. Clin. Microbiol. , 23, 43-45.
- 6. Fernandez, M.M.; Marzi, M.C.; Berguer, P.; Buryzn, D.; Langley, R.J.; Piazzon, I.; Mariuzza, R.A.; Malchiodi, E.L. (2006). Binding of natural variants of staphylococcal superantigns SEG and SEI to TCR and MHC class II molecule. Mol. Immunol. , 43, 927, 938.
- 7. Hatakka, M.; Bjorkroth, K.J.; Asplud, K.; Mäki-Petäys, N.; Korkeala, H.J. (2000). Genotypes and enterotoxicity of isolated from the hands and nasal cavities of flight-catering employees. J. Food Prot. , 11, 1487-1491.
- 8. Jarraud, S.; Peyrat, M.A.; Lim, A.; Tristan, A.; Bes, M.; Mougel, C.; Etienne, J.; Vandenesch, F.; Bonneville, M.; Lina, G. (2001). EGC, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. , 166, 669-677.
- 9. Johnson, W.M.; Tyler, S.D.; Ewan, P.; Ashton, F.E.; Pollard, D.R.; Rozee, K.R. (1991). Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Immunol, 29, 426-430.
- 10. Kloos, W.E.; Bennerman, T.L. (1994). Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev, 7, 117-40.
- 11. Kuroda, M.; Ohta, T.; Uchiyama, I.; Baba, T.; Yuzawa, H.; Kobayashi, I.; Cui, L.; Oguchi, A.; Aoki, K.; Nagai, Y.; Lian, J.; Ito, T.; Kanamori, M.; Matsumaru, H.; Maruyama, A.; Murakami, H.; Hosoyama, A.; Mizutani-Ui, Y.; Takahashi, N.K.; Sawano, T.; Inoue, R.; Kaito, C.; Sekimizu, K.; Hirakawa, H.; Kuhara, S.; Goto, S.; Yabuzaki, J.; Kanehisa, M.; Yamashita, A.; Oshima, K.; Furuya, K.; Yoshino, C.; Shiba, T.; Hattori, M.; Ogasawara, N.; Hayashi, H.; Hiramatsu, K. (2001). Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet, 357, 1225-1240.
- 12. Lamaita, H.C.; Cerqueira, M.M.O.P.; Carmo, L.S.; Santos, D.A.; Penna, M.R.; Souza, C.F.A.M. (2005). Contagem de Staphylococcus sp e detecção de enterotoxinas estafilocócicas e toxina da síndrome do choque tóxico em amostras de leite cru refrigerado. Arq. Bras. Med. Vet. Zootec., v.57, n.5, p.702-709.
- 13. Lancette, G.A.; Bennett, R.W. (2001). Staphylococcus aureus and Staphylococcal Enterotoxins. In: Downes F.P.; Ito, K. Compendium of Methods for the Microbiological Examination of Foods (pp. 387-403). Washington: APHA.
- 14. Letertre, C.; Perelle, S.; Dilasser, F.; Fach, P. (2003). Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol., 95, 38-43.
- 15. McLauchlin, J.; Narayanan, G.L.; Mithani, V.; O'Neill, G. (2000). The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. , 63, 479-488.
- 16. Mehrotra, M.; Wang, G.; Johnson, W.M. (2000). Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. , 38, 1032-1035.
- 17. Munson, S.H.; Tremaine, M.T.; Betley, M.J.; Welch, R.A. (1998). Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun., 66, 3337-3348.
- 18. Nashev, D.; Toshkova, K.; Isrina, S.; Salaisa, S.; Hassan, A.A.; Lammler, C.; Zschock, M. (2004). Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers. FEMS Microbiol. Lett. , 233, 45-52.
- 19. Normanno, G.; Firinu, A.; Virgilio, S.; Mula, G.; Dambrosio, A.; Poggiu, A.; Decastelli, L.; Mioni, R.; Sucuota, S.; Bolzoni, G.; Di Giannatale, E.; Salinetti, A.P.; La Salandra, G.; Bartoli, M.; Zuccon, F.; Pirino, T.; Sias, S.; Parisi, A.; Quaglia, N.C.; Celano, G.V. (2005). Coagulase-positive Staphylococci and Staphylococcus aureus in foods products marketed in Italy. Food Microbiol., 98, 73-79.
- 20. Omoe, K.; Ishikama, M.; Shimoda, Y.; Hu, D.L.; Ueda, S.; Shinagawa, K. (2002). Detection of seg, seh and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolate harboring seg, she or sei genes. J. Clin. Microbiol., 40, 857-862.
- 21. Omoe, K.; Hu, D.L.; Takahashi-Omoe, H.; Nakane, A.; Shinagawa, K. (2003). Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun, 71, 6088-6094.
- 22. Orwin, P.M.; Leung, D.Y.M.; Donahue, H.L.; Novick, R.P.; Schlievert, P.M. (2001). Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun, 69, 360-366.
- 23. Orwin, P.M.; Fitzgerald, J.R.; Leung, D.Y.M.; Gutierrez, J.A.; Bohach, G.A.; Schlievert, P.M. (2003). Characterization of Staphylococcus aureus enterotoxin L. Infect. Immun, 71, 2916-2919.
- 24. Peacock, S.; Moore, C.; Justice, A.; Kanlazanou, M.; Stou, L.; Mackie, K.; O'Neil, G.; Day, N. (2002). Virulent combinations of adhesin and toxin genes in natural population of Staphylococcus aureus. Infect. Immun, 70, 4987-96.
- 25. Rapini, L.S.; Cerqueira, M.M.O.P.; Carmo, L.S.; Veras, J.F.; Souza, M.R. (2005). Presença de Staphylococcus spp produtores de enterotoxinas e da toxina da síndrome do choque tóxico em manipuladores de queijo de cabra. Arq. Bras. Med. Vet. Zootec., v.57, n.6, p.825-829.
- 26. Ren, K.; Bannan, J.D.; Pancholi, V.; Cheung, A.L.; Robbins, J.C.; Fischetti, V.A.; Zabriskie, J.B. (1994). Characterization and biological properties of a new staphylococcal exotoxin. J. Exp. Med. , 180, 1675-83.
- 27. Rosec, J.P.; Gigaud, O. (2002). Staphylococcal enterotoxin genes of classical and new types detected by PCR in France. Int. J. Food Microbiol. , 77, 61-70.
- 28. Santos, D.A. (2003). O papel do manipulador de alimentos em surtos de intoxicação alimentar causados por espécies de Staphylococcus ocorridos em quatro cidades do Estado de Minas Gerais, Brasil 85f. Dissertação (Mestrado em Ciências Biológicas) - Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte.
- 29. Stanford, T.L.M.; Silva, C.G.M.; Mota, R.A. (2006). Enterotoxigenicidade de Staphylococcus spp isolados de leite in natura. Ciênc. Tecnol. Aliment., 26, 41-45.
- 30. Su, Y.C.; Wong, A.C.L. (1995). Identification and purification of a new staphylococcal enterotoxin, H. Journal of Applied and Environmental Microbiology, 61, 1438-43.
- 31. Vandenbergh, M.F.Q.; Yzermann, E.P.F.; Belkum, A.V.; Boelens, H.A.M.; Sijmons, M.; Verrugh, H.A. (1999). Follow-up of S. aureus nasal carriage after years: redefining the persistent carrier state. J. Clin. Microbiol. , 10, 3133-3140.
- 32. Veras, J.F.; Carmo, L.S.; Tong, L.C.; Shupp, J.W.; Cummings, C.; Santos, D.A.; Cerqueira, M.M.O.P.; Cantini, A.; Nicoli, J.R.; Jett, M. (2008). A study of the enterotoxigenicity of coagulasenegative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infec. Dis., 12, 410-415.
- 33. Von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. , 344, 11-6.
- 34. Zhang, S.; Iandolo, J.J.; Stewart, G.C. (1998). The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett., 168, 227-233.
Publication Dates
-
Publication in this collection
23 Nov 2009 -
Date of issue
Mar 2010
History
-
Received
08 May 2008 -
Accepted
28 Sept 2009 -
Reviewed
20 Apr 2009