Abstracts
The objective of the present study was to improve the detection of B. abortus by PCR in organs of aborted fetuses from infected cows, an important mechanism to find infected herds on the eradication phase of the program. So, different DNA extraction protocols were compared, focusing the PCR detection of B. abortus in clinical samples collected from aborted fetuses or calves born from cows challenged with the 2308 B. abortus strain. Therefore, two gold standard groups were built based on classical bacteriology, formed from: 32 lungs (17 positives), 26 spleens (11 positives), 23 livers (8 positives) and 22 bronchial lymph nodes (7 positives). All samples were submitted to three DNA extraction protocols, followed by the same amplification process with the primers B4 and B5. From the accumulated results for organ, the proportion of positives for the lungs was higher than the livers (p=0.04) or bronchial lymph nodes (p=0.004) and equal to the spleens (p=0.18). From the accumulated results for DNA extraction protocol, the proportion of positives for the Boom protocol was bigger than the PK (p<0.0001) and GT (p=0.0004). There was no difference between the PK and GT protocols (p=0.5). Some positive samples from the classical bacteriology were negative to the PCR and viceversa. Therefore, the best strategy for B. abortus detection in the organs of aborted fetuses or calves born from infected cows is the use, in parallel, of isolation by classical bacteriology and the PCR, with the DNA extraction performed by the Boom protocol.
bovine; brucellosis; abortion; PCR; DNA extraction
O objetivo do presente estudo foi aperfeiçoar a detecção de Brucella abortus pela PCR em homogeneizados de órgãos de fetos abortados por vacas infectadas, importante mecanismo para descobrir focos da doença na fase de erradicação. Assim, foram comparados diferentes protocolos de extração de DNA, visando à detecção de B. abortus pela PCR em amostras clínicas colhidas de abortos ou de bezerros oriundos de vacas desafiadas com a estirpe 2308 de B. abortus. Para tanto, foram construídos dois grupos padrão ouro com base na bacteriologia clássica, constituídos por: 32 pulmões (17 positivos), 26 baços (11 positivos), 23 fígados (8 positivos) e 22 linfonodos bronquiais (7 positivos). Todas essas amostras foram submetidas a três protocolos de extração de DNA, seguidos do mesmo processo de amplificação com os primers B4 e B5. Nos resultados acumulados por órgão, a proporção de positivos nos pulmões foi maior que a encontrada nos fígados (p=0,04) e nos linfonodos bronquiais (p=0,004), e igual a verificada nos baços (p=0,18). Nos resultados acumulados por método de extração de DNA, a proporção de positivos para o protocolo de Boom foi maior que a verificada para o PK (p<0,0001) e GT (p=0,0004). Não houve diferença entre os protocolos PK e GT (p=0,5). Algumas amostras positivas ao isolamento foram negativas à PCR e vice-versa. Assim, a melhor estratégia para se pesquisar B. abortus em tecidos de fetos abortados ou de bezerros nascidos de vacas infectadas é a utilização, em paralelo, do isolamento e da PCR, com extração do DNA pelo método do Boom.
brucelose; bovinos; aborto; PCR; extração DNA
VETERINARY MICROBIOLOGY
Evaluation of DNA extraction protocols for Brucella abortus pcr detection in aborted fetuses or calves born from cows experimentally infected with strain 2308
Avaliação de diferentes protocolos de extração de DNA para detecção de Brucella abortus a partir de abortos ou de bezerros nascidos de vacas experimentalmente infectadas com estirpe 2308
Matrone, M.; Keid, L.B.; Rocha, V.C.M.; Vejarano, M.P.; Ikuta, C.Y.; Rodriguez, C.A.R.; Ferreira, F.; Dias, R.A.; Ferreira Neto, J.S* * Corresponding Author. Mailing address: Departamento de Medicina Veterinária Preventiva e Saúde Animal, Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo - Av. Prof. Dr. Orlando Marques de Paiva, 87, Cidade Universitária, CEP 05508-270, São Paulo, SP, Brasil. Email: jsoares@vps.fmvz.usp.br
Departamento de Medicina Veterinária Preventiva e Saúde Animal, Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo, São Paulo, SP, Brasil
ABSTRACT
The objective of the present study was to improve the detection of B. abortus by PCR in organs of aborted fetuses from infected cows, an important mechanism to find infected herds on the eradication phase of the program. So, different DNA extraction protocols were compared, focusing the PCR detection of B. abortus in clinical samples collected from aborted fetuses or calves born from cows challenged with the 2308 B. abortus strain. Therefore, two gold standard groups were built based on classical bacteriology, formed from: 32 lungs (17 positives), 26 spleens (11 positives), 23 livers (8 positives) and 22 bronchial lymph nodes (7 positives). All samples were submitted to three DNA extraction protocols, followed by the same amplification process with the primers B4 and B5. From the accumulated results for organ, the proportion of positives for the lungs was higher than the livers (p=0.04) or bronchial lymph nodes (p=0.004) and equal to the spleens (p=0.18). From the accumulated results for DNA extraction protocol, the proportion of positives for the Boom protocol was bigger than the PK (p<0.0001) and GT (p=0.0004). There was no difference between the PK and GT protocols (p=0.5). Some positive samples from the classical bacteriology were negative to the PCR and viceversa. Therefore, the best strategy for B. abortus detection in the organs of aborted fetuses or calves born from infected cows is the use, in parallel, of isolation by classical bacteriology and the PCR, with the DNA extraction performed by the Boom protocol.
Key words: bovine, brucellosis, abortion, PCR, DNA extraction
RESUMO
O objetivo do presente estudo foi aperfeiçoar a detecção de Brucella abortus pela PCR em homogeneizados de órgãos de fetos abortados por vacas infectadas, importante mecanismo para descobrir focos da doença na fase de erradicação. Assim, foram comparados diferentes protocolos de extração de DNA, visando à detecção de B. abortus pela PCR em amostras clínicas colhidas de abortos ou de bezerros oriundos de vacas desafiadas com a estirpe 2308 de B. abortus. Para tanto, foram construídos dois grupos padrão ouro com base na bacteriologia clássica, constituídos por: 32 pulmões (17 positivos), 26 baços (11 positivos), 23 fígados (8 positivos) e 22 linfonodos bronquiais (7 positivos). Todas essas amostras foram submetidas a três protocolos de extração de DNA, seguidos do mesmo processo de amplificação com os primers B4 e B5. Nos resultados acumulados por órgão, a proporção de positivos nos pulmões foi maior que a encontrada nos fígados (p=0,04) e nos linfonodos bronquiais (p=0,004), e igual a verificada nos baços (p=0,18). Nos resultados acumulados por método de extração de DNA, a proporção de positivos para o protocolo de Boom foi maior que a verificada para o PK (p<0,0001) e GT (p=0,0004). Não houve diferença entre os protocolos PK e GT (p=0,5). Algumas amostras positivas ao isolamento foram negativas à PCR e vice-versa. Assim, a melhor estratégia para se pesquisar B. abortus em tecidos de fetos abortados ou de bezerros nascidos de vacas infectadas é a utilização, em paralelo, do isolamento e da PCR, com extração do DNA pelo método do Boom.
Palavras-chave: brucelose, bovinos, aborto, PCR, extração DNA
INTRODUCTION
Brucellosis, caused by Brucella abortus, is one of the most wide-spread zoonosis in the world (3). In bovines the disease is related to reproductive problems, with reduction of meat and milk production, while in humans it causes unspecific symptoms of generalized infection (18).
Classically, direct diagnosis is performed by cultivation in artificial media, with posterior identification of the isolates by its morphology and growth characteristics of the colonies, biochemical tests and phagotyping (18). Disadvantages of these procedures are the high costs, time necessary for growth and identification of the isolates, apart from high risk for personnel (10).
An alternative to the classic bacteriological methods is the Polimerase Chain Reaction (PCR), where a specific fragment of the bacterial DNA present in the sample is detected (18). However, few studies have been performed with field samples to evaluate this diagnostic method for brucellosis (13).
Although PCR is a diagnostic procedure that is known to have advantages of being fast, highly specific and sensitive (4, 9, 14), in some cases it may show low sensitivity due to the presence of factors inhibiting amplification of the DNA, like serum proteins, somatic cell debris, polysaccharides and other components of body tissues and fluids (23). These inhibiting factors are the main cause of false negative results.
In the context of a surveillance system for bovine brucellosis, one possibility of detecting infected herds is via bacteriological analysis of aborted fetuses (18). The best samples for isolation of Brucella spp from bovine fetuses are abomasal liquid, rectal swab, bronchial lymph nodes, lung, liver and spleen (19). These samples, especially organ homogenates, are subject to inhibitors of DNA amplification (23). The solution for this problem, diminishing the number of false negative results to PCR, can be the protocol for DNA extraction.
There are some promising DNA extraction protocols for B. abortus PCR diagnosis in organs. The method that uses proteinase K (pK) to promote the cellular lysis, proposed to detect B. mellitensis in blood samples of goats (15), showed a good sensitivity to diagnose brucellosis in samples of bovine abortions artificially contaminated (8). The protocol that uses guanidine-isotiocianate (GT) to promote the cellular lysis and the nucleic acids purification is practical, fast and allows simultaneous isolation of RNA, DNA and proteins from cells and tissue samples (7). The method proposed by Boom and colleagues (2), also based on the use of guanidineisotiocianate, but combined with the chelanting action of diatomaceous earth, is simple, fast and reliable to the purification of DNA and RNA from human serum and urine (2), presenting good results in the detection of Mycobacterium bovis in homogenates of tissue samples with tuberculous lesions obtained from bovine slaughterhouses (20).
Thus, bearing in mind the importance of bacteriological analysis of aborted fetuses for a brucellosis surveillance system and the occurrence of false negative results in PCR, when the clinical samples consist of homogenized organs, the present study compared three protocols for DNA extraction from homogenized organ samples.
MATERIAL AND METHODS
Samples and experimental design
The positive and negative gold standard groups were composed of homogenates of organ fragments originating from a clinical test of vaccine against brucellosis conducted in the Laboratory of Bacterial Zoonoses (LZB) of the Faculty of Veterinary Medicine and Zootechny of the University of São Paulo, Brazil. All samples came from aborted fetuses or calves, born from cows infected with the strain 2308 of B. abortus by approximately the fifth month of pregnancy, with a dose of 3.5 x 107 bacteria in the conjunctive sac. The positive gold standard group was composed of 17 samples of lungs, 11 of spleens, 8 of livers, and 7 of bronchial lymph nodes, from which B. abortus 2308 was isolated by classic bacteriological methods. The negative gold standard group was composed of 15 samples each of these organs, where classic bacteriological isolation of brucella was not possible.
All samples were submitted to three different DNA extraction protocols and subsequently to the same methods for amplification and revelation. The volume of homogenates used for DNA extraction protocols was consistently 400 µL.
Tests with pure cultures
A suspension of the strain 2308 of B. abortus diluted in sterile saline solution was prepared and standardized by its turbidity corresponding to 0.5 on the McFarland scale (0.5 mL BaCl2.2H2O 0,048M - 1.75% by weight/volume in 99.5 mL H2SO4 0.36 N - 1% volume/volume), corresponding to a concentration of 1.5 x 108 bacteria/mL. From this suspension, a series of 13 further ten-fold dilutions was obtained. Of each of the 14 dilutions, 100 µL were inoculated in triptose agar for counting the number of Colony Forming Units (CFU), and 400 µL were used for each of the three DNA extraction protocols tested in this study. All samples of extracted DNA were submitted to the same method for amplification and revelation.
DNA extraction protocols
1) Lysis with proteinase K (pK) and phenol/chlorophorm (adapted from Leal et al. (15)): Add 1000 µL of TE, 400 µL of the sample and vortex for 10 seconds. Centrifuge at 13000 X g for 5 minutes and remove the supernatant (repeat until suspension becomes clear). Resuspend sediment in 300 µL of lysis buffer [10 µL of pK 20 units/mg, 50 µL of 10 % sodium dodecyl sulfate (SDS), 5 µL Tris-HCL 1M, 25 µL EDTA 0.5 M, 10 µL NaCL 5M and 400 µL of ultrapure water]. Incubate in dry bath at 37º C/overnight at 600 rpm. Add 500 µL of phenol, vortex for 20 seconds and centrifuge at 13000 X g for 5 minutes. Transfer 200 µL of the aqueous phase to a new tube, carefully avoiding to pipet the organic interphase. Add 300 µL of phenol/chlorophorm, vortex for 10 seconds and centrifuge at 13000 X g for 5 minutes. Transfer 200 µL of the aqueous phase to a new tube, carefully avoiding to pipet the organic interface. Add 200 µL of propane and homogenize by inversion. Maintain at -20º C for 4 hours. Centrifuge at 13000 X g for 30 minutes, remove the supernatant and add 500 µL of 70% ethanol. Centrifuge at 13000 X g for 30 minutes and remove supernatant, letting sample dry at room temperature. Add 30 µL of TE pH 8.0, incubate in a dry bath at 56º C /15 minutes and store at -20º C.
2) Lysis with guanidine isothiocyanate GT (adapted from Chomkzynski (7)): Add 900 µL of GT (60g of guanidine isothiocyanate + 5 mL of Tris-HCl 1M pH 7.5 + 10 mL of EDTA 0.25 M pH 8.0 + 100 mL of phenol + ddH2O qsp 100 mL) to 400 µL of the sample and vortex for 15 seconds. Wait 10 minutes and vortex again. Add 200 µL of chlorophorm, vortex for 15 seconds and let rest for 10 minutes. Centrifuge at 12000 X g for 5 minutes, recuperate supernatant (700µL) and add equal volume of propanol for DNA precipitation. Homogenize manually and freeze at -20º C for 2 hours. Centrifuge at 12000 X g for 20 minutes and carefully dispose of the supernatant. Add 500 µL of 70% ethanol and homogenize manually. Centrifuge at 12000 X g for 10 minutes, remove supernatant and let dry at room temperature. Add 30 µL of TE, vortex for 10 seconds and put in dry bath at 56º C for 15 minutes. Store frozen at -20º C.
3) Lysis with guanidine isothiocyanate followed by treatment with carrier suspension (diatomaceous earth) (Boom) (adapted from Boom et al. (2)): Add 1000 µL of lysis buffer (120 g GuSCN - 10 mL of 1 M Tris-HCL pH 6.4 - 8.8 mL of EDTA 0.5 M pH 8.0 - 1 mL Triton x100) to 400 µL of the sample and vortex for 2 minutes. Add 40 µL of the carrier suspension (1 g of diatomaceous earth - 50 µL of HCL 37% - 5 mL of ultrapure water), vortex for 1 minute and let rest for 20 minutes. Centrifuge at 12800 X g for 2 minutes at 4º C and dispose of the supernatant. Add 500 µL of washing buffer to the pellet (120g of GuSCN - 10 mL of 1M Tris-HCL pH 6.4 e 100 mL of ultrapure water), vortex for 1 minute and centrifuge at 12800 X g for 2 minutes at 4º C. Dispose of the supernatant and repeat the washing buffer procedure. Add 500 µl of 70% ethanol, vortex for 1 minute and centrifuge at 12800 X g for 2 minutes at 4º C. Dispose of the supernatant and repeat the procedure with 70% ethanol. Add 500 µl of acetone, vortex for 1 minute and centrifuge at 12800 X g for 2.5 minutes at 4º C. Dispose of the supernatant and place the open tube in an incubator at 37º C for at least 30 minutes. Add 150 µL of eluition buffer (1 mL of Tris-HCL 1M - 0.2 mL of EDTA pH 8.0 and 98.8 mL of ultrapure water), vortex for 1 minute and incubate in a dry bath at 55º C for 10 minutes. Vortex for 1 minute and centrifuge at 12800 X g for 5 minutes. Transfer 90 µL of the aqueous phase to a new tube and centrifuge at 12800 X g for 5 minutes. Store at -20º C.
Amplification and revelation
The primers used were B4 (5´ TGGCTCGGTTGCCAATATCAA 3´) and B5 (3´ CGCGCTTGCCTTTCAGGTCTG 5´), described by Baily et al. (1), which amplify a fragment of 223 bp. PCR was realized in a volume of 50 µL, consisting of 22.5 µL of ultrapure water, 5 µL of reaction buffer 10 X (500 nM KCL; 15 nM MgCl2, 100 nM TrisHCL, pH 9.0), 8.0 µL of dNTP mixture (200 nM of each nucleotide [dCTP, dATP, dGTP, dTTP]), 1.5 µL of MgCl2 (50 nM), 5 µL of the primer B4 (10 pmol/ µL), 5 µL of the primer B5 (10 pmol/ µL), 0.5 µL of Taq DNA polimerase (5 units per µL), and 2.5 µL of the DNA sample extracted using pK, 2.5 µL of the DNA sample in a 1:5 TE dilution following the GT protocol, or 5 µL of the DNA sample extracted following Boom (the quantity of water was regulated according to the quantity of DNA used for each extraction protocol). Amplification was realized adopting an initial denaturation of 94º C for 5 minutes, and 40 cycles of 94º C for 1 minute (denaturation), 60º C for 1 minute (annealing) and 72º C for 1 minute (extension) with a final extension of 72º C for 10 minutes (1). Finally, all amplification products were analyzed using electrophoresis in an agarose gel containing 1.5% ethidium bromide at 0.5µg/mL and subsequent observation using an ultraviolet transilluminator.
Data Analysis
For each of the protocols, relative sensitivity and its confidence interval were calculated considering isolation or experimental inoculation as gold standard. Proportions were compared using a Chisquare test. Calculations were carried out with the Epi Info 6.0 and Med Calc 8.2 programs.
RESULTS
Table 1 presents the results from the tests conducted with pure B. abortus culture, showing the thresholds of isolation by classic bacteriological method and detection by PCR, performed with samples from different DNA extraction protocols.
The dilutions with concentrations from 1.5 x 108 and 1.5 x 104 bacteria/mL showed uncountable numbers of CFU in the quantitative cultures. The samples with 1.5 x 103, 1.5 x 102 and 1.5 x 101 bacteria/mL showed 4200, 240 and 50 UFC/mL, respectively. The samples with 1.5 x 100 to 1.5 x 105 bacteria/mL showed no growth.
Table 2 shows results from PCR following the three DNA extraction protocols, compared with those obtained for isolation using classical bacteriology, for the organ samples. Table 3 shows values of sensitivity for the DNA extraction protocols based on organ samples, considering the results from classic bacteriology as gold standard. Both tables show accumulated results for DNA extraction protocols and organs.
Considering Table 2 data, the accumulated results for DNA extraction protocols show no statistical difference among them, concerning the proportions of positive PCR results. Analyzing the same Table data, the accumulated results for organs show that the proportion of positive PCR results for spleen was significantly higher than the bronchial lymph node (p=0.01) or liver (p=0.05), and equal to the lung (p=0.25).
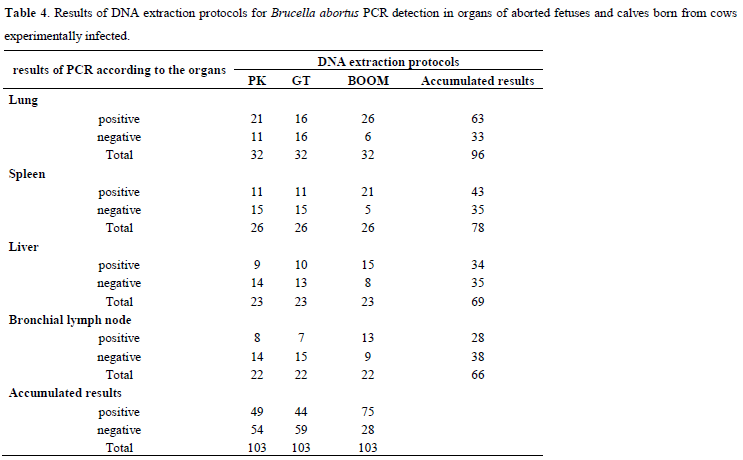
All fetuses and calves sampled were born from cows experimentally infected with the strain 2308 of B. abortus as part of a clinical test of vaccine against brucellosis. Thus, considering the experimental infection as gold standard, Tables 4 and 5 were elaborated. Table 4 shows the results from PCR for the three DNA extraction protocols and Table 5 presents the values of relative sensitivity.
Considering Table 4 data, the accumulated results for organ show that the proportion of PCR positive results for the lungs was significantly higher than the livers (p=0.04) or bronchial lymph nodes (p=0.004), and equal to the spleens (p=0.18). Analyzing the same Table data, the accumulated results for DNA extraction protocol show that the proportion of positive PCR results for the Boom protocol was significantly higher than pK (p<0.0001) and GT (p=0.0004). There was no difference between the pK and GT protocols (p=0.5).
DISCUSSION
The lowest threshold concentration for detection of B. abortus was verified for the pK protocol (1.5 10-2 bacteria/mL), followed by Boom (1.5 x 101 bacteria/mL) and GT (1.5 x 105 bacteria/mL) (Table 1). These results indicate that the pK protocol detected B. abortus in a dilution containing 0.015 bacteria/mL, which is not feasible. This was probably the consequence of the low precision of quantification using the McFarland scale. In fact, the CFU count obtained from the bacterial suspensions showed a number always higher than expected from the scale. Apart from that, it is reasonable to suppose that there are CFU produced by more than one bacterial cell. Thus, the McFarland scale and the CFU counts under-quantified the number of bacilli present in the suspensions.
Considering classic bacteriological isolation as gold standard, the relative sensitivity of PCR reached 100% for any tested DNA extraction protocols only for spleen samples (Table 3). For the organs, the highest value of relative sensitivity was found for the spleen, followed by the lung, liver, and bronchial lymph node (Table 3). The accumulated results for organs, presented in Table 2, show that the proportion of positive PCR results for spleen was significantly higher than the bronchial lymph node (p=0.01) or liver (p=0.05), and equal to the lung (p=0.25). When the experimental infection is considered as gold standard, the highest value of relative sensitivity was observed for the lung, followed by the spleen, liver, and bronchial lymph node (Table 5). The accumulated results for organs, presented in Table 4, show that the proportion of PCR positive results for the lungs was significantly higher than the livers (p=0.04) or bronchial lymph nodes (p=0.004), and equal to the spleens (p=0.18). Thus, the best organs to detect B. abortus using PCR in aborted fetuses or calves born from infected cows were lung and spleen.
Considering isolation as gold standard, the pK DNA extraction protocol presented the highest value of relative sensitivity, followed by Boom and GT (Table 3). However, there were no statistically significant differences among proportions of positive PCR results. Cortez et al. (8), stated that the pK protocol was effective in extracting DNA of B. abortus. Interestingly, some of the 43 samples with positive results for isolation showed negative results for PCR: 2 for pK, 6 for GT and 5 for Boom (Table 2). When the experimental infection is considered as gold standard, the Boom DNA extraction protocol presented the highest value for relative sensitivity, followed by pK and GT (Table 5). The accumulated results for DNA extraction protocol, presented in Table 4, show that the proportion of positive PCR results for the Boom protocol was significantly higher than pK (p<0.0001) and GT (p=0.0004). There was no difference between the pK and GT protocols (p=0.5).
Ribeiro (20) compared three DNA extraction protocols for detection of Mycobacterium bovis in homogenized organ samples and demonstrated that the Boom protocol was the best. The lower sensitivity of the GT protocol could have been the result of the presence of a large quantity of factors inhibiting amplification of the nucleic acid, like serum proteins, somatic cell debris, polysaccharides and other components of body fluids (23), or large quantity of host DNA, as this method does not include pre-washing of samples, a procedure that was part of the pK protocol, and performed as an intermediary step in the Boom protocol. In the 60 samples where isolation of B. abortus was not possible, PCR showed positive results in 8 samples for the pK protocol, 6 for GT, and 37 for Boom (Table 2). For these samples, the proportion of positive PCR found for the Boom extraction was significantly higher than pK (p=0.005) and GT (p=0.001). As all samples used in this study came from aborted fetuses or calves born from experimentally infected cows, it is reasonable to suppose the PCR results are not false positives, although isolation showed a negative result. In studies about detection of B. abortus in samples from bovine abortions, Cortez et al. (8), detected 4 positive samples using PCR in 54 samples classified as negative by classic isolation. Fekete et al. (11), obtained 2 positive samples in 52 negatives for microbiologic cultivation.
The specificity of the DNA extraction protocols for PCR was not calculated because it depends on the primers used only. In the present experiment the primers B4 and B5, described by Baily et al. (1) for detection of B. abortus and B. mellitensis, were used. The authors compared these primers with the nucleotide sequence databases Genbank and EMBL (European Molecular Biology Laboratory) and did not identify any significant sequences homology with other bacteria. These primers have been used with success to diagnose infection with brucella by various authors (8, 17, 21, 22,), none of whom ever related false positives. Unspecificity was only reported for human infection with Ochrobactrum anthropi (5, 6, 12, 16).
For B. abortus PCR detection in homogenized organ samples from aborted fetuses or calves born from infected cows, these results allow concluding that: 1) the lung and spleen presented higher probability of success than liver and bronchial lymph node, 2) the DNA extraction protocol influences the sensitivity of the PCR and the best one was the socalled Boom protocol.
Thus, in spite of the disadvantages of the classical bacteriologic methods, the best strategy to investigate B. abortus in tissues of aborted fetuses or born calves of infected cows is the parallel use of isolation and PCR, using the Boom protocol for the DNA extraction.
ACKNOWLEDGEMENTS
Thanks to CAPES for a master scholarship and FAPESP for financial support.
Submitted: October 21, 2008;
Returned to authors for corrections: February 16, 2009;
Approved: May 04, 2009.
- 1. Baily, G.G.; Krahn, J.B.; Drasar, B.S.; Soker, N.G. (1992). Detection of Brucella melitensis and Brucella abortus by DNA amplification. J. Trop. Med. Hyg 95, 271-275.
- 2. Boom, R.; Sol, C.J.A.; Salimans, M.M.M.; Jansen, C.L.; WertheimVan Dillen, P. M.E.; Van Der Noordaa, J. (1990). Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol 28 (3), 495-503.
-
3Brasil, Ministério da Agricultura, Pecuária e Abastecimento (MAPA). Planos e Programas. Programas. Área Animal. (PNCEBT). Instrução Normativa Ministerial 2/2001 institui o Programa Nacional de Controle e Erradicação da Brucelose e da Tuberculose (PNCEBT). Disponível em: www.agricultura.gov.br/pls/portal/docs/PAGE/MAPA/PROGRAMAS/al.AREA_ANIMAL/ PNCEBT/INSTRUCAONORMATIVA N.PDF Acesso em: 4 de setembro de 2008.
- 4. Bricker, B.J. (2002). PCR as a diagnostic tool for brucellosis. Vet. Microbiol. 90, 435-446.
- 5. Casanas, M.C.; Queipo-Ortuno, M.I.; Rodriguez-Torres, A.; Orduna, A.; Colmenero, J.D.; Morata, P. (2001). Specificity of a polymerase chain reaction assay of a target sequence on the 31Kilodalton Brucella antigen DNA used to diagnose human brucellosis. Eur. J. Clin. Microbiol. Infect. Dis 20 (2), 127-131.
- 6. Cieslak, T.J.; Robb, M.L.; Drabick, C.J.; Fischer, G.W. (1992). Catheter-associated sepsis caused by Ochrobactrum anthropi: report of a case and review of related nonfermentative bacteria. Clin. Infect. Dis 14 (4), 902-907.
- 7. Chomkzynski, P.A. (1993). Reagent for the single step simultaneous isolation of RNA, DNA and proteins from cells and tissue samples. Biotechniques 15, 532-537.
- 8. Cortez, A.; Scarcelli, E.; Soares, R.M.; Heinemann, M.B.; Sakamoto, S.M.; Genovez, M.E.; Ferreira, F.; Richtzenhaim, L.J. (2001). Detection of Brucella DNA from aborted bovine foetuses by polymerase chain reaction. Aust. Vet. J 79 (7), 500-501.
- 9. Dahouk, S.A.; Tomaso, H.; Nockler, K.; Neubauer, H.; Frangoulidis, D. (2003). Laboratory based diagnosis of brucellosisA review of the literature. Clin. Lab 49, 487-505.
- 10. Fekete, A.; Bantle, J.A.; Halling, S.M. (1992b). Detection of Brucella by polymerase chain reaction in bovine fetal and maternal tissues. J. Vet. Invest. 4, 79-83
- 11. Fekete, A.; Bantle, A.J.; Halling, S.M. (1990a). Preliminary development of diagnostic test for Brucella using polymerase chain reaction. J. Appl. Bacteriol 69, 216-227.
- 12. Gransden, W.R.; Eykyn S.J. (1992). Seven cases of bacteremia due to Ochrobactrum anthropi. Clin. Infect. Dis., 15 (6), 1068-1069.
- 13. Guler, L.; Gunduz, K.; OK, U. (2003). Comparison of polymerase chain reaction and bacteriological culture for the diagnosis of sheep brucellosis using aborted fetus samples. Vet. Microbiol., v. 93, p. 53-61, 2003.
- 14. Lage, A.P.; Roxo, E.; Muller, E.; Poester, F.; Cavalléro, J.C.M.; Ferreira Neto, J.S.; Mota, P.M.P.C.; Gonçalves, V.S.P. (2006). Programa nacional de controle e erradicação da brucelose e da tuberculose animal (PNCEBT) Brasília, Ministério da Agricultura, Pecuária e Abastecimento.
- 15. Leal-Klevezas, D.S.; Martinez-Vazquez, I.O.; Lopez-Merino, L.; Martinez-Soriano, J.P. (1995a). Single-stpe PCR for detection of Brucella spp From blood and milk of infected animals. J. Clin. Microbiol 12, 3087-3090.
- 16. Manfredi, R.; Nanetti, A.; Ferri, M.; Chiodo, F. (1999). Nosocomial sepsis due to Ochrobactrum anthropi in HIV positive patients: two case reports. Infez. Med. 7 (2), 119-124.
- 17. Matar, G.M.; Khneisser, I.A.; Abdelnoor, A.M. (1996). Rapid laboratory confirmation of human brucellosis by PCR analysis of a target sequence on the 31 kilodalton Brucella antigen DNA. J. Vet. Microbiol. 34 (2), 477-478.
- 18. Paulin, L.M.; Ferreira Neto, J.S. (2003). O combate à brucelose bovina Jaboticabal, FUNEP.
- 19. Poester, F.P.; Gonçalves, V.S.; Paixao, T.A.; Santos, R.L.; Olsen, S.C.; Schurig, G.G.; Lage, A.P. (2006). Efficacy of strain RB51 vaccine in heifers against experimental brucellosis. Vaccine 24 (25), 5327-5534.
- 20. Ribeiro, D.C. (2006). Comparação de protocolos de extração de DNA para detecção de Mycobacterium bovis através da PCR em homogeneizado de órgãos São Paulo, Brasil, 55p. (M. Sc. Dissertation. Faculdade de Medicina Veterinária. USP)
- 21. Richtzenhain, L.J.; Cortez, A.; Heinemann, M.B.; Soares, R.M.; Sakamoto, S.M.; Vasconcellos, S.A.; Higa, Z.M.M.; Scarcelli, E.; Genovez, M.E. (2002). A multiplex PCR for the detection of Brucella spp and Leptospira spp DNA from aborted bovine fetuses. Vet. Microbiol 87, 139-147.
- 22. Zerva, L.; Bourantas, K.; Mitka, S.; Kansouzidou, A.; Legakis, N.J. (2001). Serum is the preferred clinical specimen for diagnosis of human brucellosis by PCR. J. Clin. Microbiol 39 (4), 1661-1664.
- 23. Wilson, I.G. (1997). Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol, 3741-3751.
Publication Dates
-
Publication in this collection
19 Aug 2009 -
Date of issue
Sept 2009
History
-
Received
21 Oct 2008 -
Reviewed
16 Feb 2009 -
Accepted
04 May 2009