Abstracts
The detection of Streptococcus mutans isolates with high genetic variability in different individuals indicates the occurrence of transmissibility. In this context, nine low-income families (40 individuals in total) with similar social conditions were assessed to identify the bioserotypes of S. mutans using both biochemical and RAPD markers and to establish the degree of similarity among the intra-familial isolates. The polymorphism analysis used the coefficient of Jaccard in both a Principal Coordinates Analysis (PCO) and in a UPGMA clustering method. A total of 157 isolates were obtained from salivary samples, using the morphology of colonies recovered using MSB agar as indicators. From those, 64 were characterized biochemically as S. mutans and 10 as S. sombrinus. Genetic variability among isolates based on RAPD markers was not consistent with their intra-familial distribution. In particular, some individuals might have experienced multiple infections given the high genetic variability among their isolates. The occurrence of 4 isolates with 100% genetic similarity is indicative of intra-familial transmission.
Streptococcus mutans; genetic variability; RAPD
A detecção de isolados de S. mutans com alta similaridade genética em indivíduos diferentes, sugere a existência de transmissibilidade. Neste contexto, nove famílias (40 indivíduos) de baixo poder aquisitivo com condições sociais homogêneas foram avaliadas, visando identificar os biosorotipos de S. mutans por meio de bioquimismo e marcadores RAPD e estabelecer o grau de similaridade entre os isolados intra-familiar. Para a análise de polimorfismo utilizou-se coeficiente de Jaccard, análise de coordenadas principais (PCO) e método "UPGMA". Foram obtidos 157 isolados, a partir de amostras salivares, usando como indicador a morfologia das colônias recuperadas em ágar MSB. Destes, 64 foram caracterizados bioquimicamente como S. mutans e 10 como S. sobrinus. A partir dos marcadores RAPD verificou-se variabilidade genética entre isolados, sendo diferente a distribuição intra-familiar destes. Em determinados indivíduos podem ter ocorrido infecções múltiplas, devido a grande variabilidade dos isolados. A existência de quatro isolados apresentando 100% de similaridade, sugeriu transmissão intra-familiar.
Streptococcus mutans; variabilidade genética; RAPD
GENERAL MICROBIOLOGY
Genetic variability of Streptococcus mutans isolated from low-income families, as shown by RAPD markers
Variabilidade genética de Streptococcus mutans isolados de famílias de baixa renda, demonstrados por marcadores RAPD
Mônica MoreiraI; Vânia Aparecida VicenteI* * Corresponding Author. Mailing address: Programa de Mestrado em Microbiologia, Parasitologia e Patologia, Departamento de Patologia Básica, Setor de Ciências Biológicas; Centro Politécnico, Universidade Federal do Paraná UFPR; Jardim das Américas, CEP 81531-990, Curitiba, PR, Brasil. Tel.: (+5541) 3356-1353. Fax: (+5541) 3266-2042. E-mail: vicente@ufpr.br ; Chirlei GlienkeII
IDepartamento de Patologia Básica, Universidade Federal do Paraná, Curitiba, PR, Brasil
IIDepartamento de Genética, Setor de Ciências Biológicas, Universidade Federal do Paraná, Curitiba, PR, Brasil
ABSTRACT
The detection of Streptococcus mutans isolates with high genetic variability in different individuals indicates the occurrence of transmissibility. In this context, nine low-income families (40 individuals in total) with similar social conditions were assessed to identify the bioserotypes of S. mutans using both biochemical and RAPD markers and to establish the degree of similarity among the intra-familial isolates. The polymorphism analysis used the coefficient of Jaccard in both a Principal Coordinates Analysis (PCO) and in a UPGMA clustering method. A total of 157 isolates were obtained from salivary samples, using the morphology of colonies recovered using MSB agar as indicators. From those, 64 were characterized biochemically as S. mutans and 10 as S. sombrinus. Genetic variability among isolates based on RAPD markers was not consistent with their intra-familial distribution. In particular, some individuals might have experienced multiple infections given the high genetic variability among their isolates. The occurrence of 4 isolates with 100% genetic similarity is indicative of intra-familial transmission.
Key-words: Streptococcus mutans, genetic variability, RAPD
RESUMO
A detecção de isolados de S. mutans com alta similaridade genética em indivíduos diferentes, sugere a existência de transmissibilidade. Neste contexto, nove famílias (40 indivíduos) de baixo poder aquisitivo com condições sociais homogêneas foram avaliadas, visando identificar os biosorotipos de S. mutans por meio de bioquimismo e marcadores RAPD e estabelecer o grau de similaridade entre os isolados intra-familiar. Para a análise de polimorfismo utilizou-se coeficiente de Jaccard, análise de coordenadas principais (PCO) e método "UPGMA". Foram obtidos 157 isolados, a partir de amostras salivares, usando como indicador a morfologia das colônias recuperadas em ágar MSB. Destes, 64 foram caracterizados bioquimicamente como S. mutans e 10 como S. sobrinus. A partir dos marcadores RAPD verificou-se variabilidade genética entre isolados, sendo diferente a distribuição intra-familiar destes. Em determinados indivíduos podem ter ocorrido infecções múltiplas, devido a grande variabilidade dos isolados. A existência de quatro isolados apresentando 100% de similaridade, sugeriu transmissão intra-familiar.
Palavras-chave: Streptococcus mutans, variabilidade genética, RAPD
INTRODUCTION
Brazil has considerably advanced over the past decade in the implementation of prevention policies for the control of caries in children. However, the situation in teenagers, adults, and elderly are among the worst in the world (28). In addition, there is still great need for dental care, even in the case of children. New governmental policies have been created to revert this situation, including several initiatives for oral health aimed at citizens of all ages, prioritizing low-income families. Therefore, the control of the disease in families needs to incorporate its microbiological, physiological, immunological, social, and cultural aspects. The multifatorial character of the disease can explain this complexity, in particular its microbiological component, which is investigated intra-familiarly worldwide (4,10,23,34).
Streptococcus mutans has been the bacterium most often associated with caries, given that this species has evolved physiological and morphological characteristics that have determined its cariogenicity and virulence (20). In addition to several structures that facilitate its adherence, such as glycocalix and fimbriae, S. mutans has enzymes that are able to convert sugar molecules into extracellular polymers (e.g., glycosyltransferase and fructosyltransferase). These polymers are responsible for the adherence of this microorganism to dental surfaces, as well as for its aggregation with other microorganisms in the oral cavity.
Studies have demonstrated that S. mutans is associated with caries in humans, and that there is a correlation between the salivary concentration of this bacterium and the risk of developing this disease. However, S. mutans is widespread, not only among populations with moderate to high incidence of the disease, but also in low-incidence populations, suggesting that the colonization by these organisms alone does not necessarily lead to the development of dental caries (2).
While seeking to identify the etiology of caries, studies have demonstrated the existence of difference genotypes of S. mutans in the mouth (11,29). In addition, several markers based on the PCR method have been utilized to identify bioserotypes of the mutans group and their relative occurrence within the oral microbiota (14,16,21,25,31,32). Among those, RAPD markers have been widely used in the study of the genetic polymorphism and in the distribution of populations of S. mutans (13,18,19,29,34,38).
Several authors have suggested different patterns in the acquisition and transmission of S. mutans based on its intra-familial distribution. Alternative hypotheses suggest that the mother is the strongest source of infection (3,4,5,22,23), or that transmission and acquisition occurs among adults (1,27,36). In addition, the role of the father as a route of transmission has also been emphasized (10,35,36), as well as other sources of transmission outside the family (7,17).
In this context, the goal of the present study is to investigate the genetic variability of S. mutans isolates using RAPD markers and their potential pattern of transmission in low-income families in the Municipality of Curitiba, PR, Brazil.
MATERIALS AND METHODS
Samples
Strains of S. mutans and S. sobrinus, were isolated from salivary samples of individuals belonging to nine families living in an industrial area at Curitiba, State of Paraná, Brazil. The studied families have a monthly per capita income between US$15 and US$50. The parents in those families had between 1 and 8 years of formal education. Reference strains of S. mutans (ATCC 25175) and S. sombrinus (ATCC 33478) were used as positive controls, and two strains of S. pyogenes (ATCC 13540 and BAA572) were used as the outgroup in the analyses.
Isolation and characterization of the isolates
Colonies were selected in MSBS agar (Mitis-Salivarius-Bacitracin-Saccarose) (12) and tested biochemically for manitol and sorbitol fermentation, arginine and esculin hydrolysis, and the production of the catalase enzyme.
DNA extraction
Samples were inoculated in BHI (Brain Heart Infusion) broth and incubated for 24 h at 36ºC. The cultures were then centrifuged at 49,000 x g for 2 min. DNA extraction was carried out using the CTAB (hexadecyl trimethyl ammonium bromide) buffer, together with a 2:1 silica gel and celite mixture under ultra sound (3 pulses of 30 s) in a modification of the protocol of De Hoog, et al. (8). DNA integrity was assessed using 0.8% gel electrophoresis, followed by visualization using ethidium bromide under UV light.
DNA amplification
The protocol for DNA amplification was based on Pereira et al. (33), with modifications. Each 15 mL reaction included 1.5 U of Taq DNA polymerase (Invitrogen), 0.4 mM of each dNTP, 4.0 mM of MgCl2, 0.4 mM of primer, 1X reaction buffer and 12 ng of template DNA. The following primers were used: OPA 2 (5' TGCCGAGCTG 3'), OPA 5 (5' AGGGGTCTTG 3'), OPA 7 (5' AAAACGGGTG 3'), OPA 9 (5' GGGTAACGCC 3'), and OPA 13 (5' CAGCACCCAC 3') (Operon Technologies).
Thermo cycling conditions followed a modification of the settings used by Spolidorio et al. (37) in a PTC-100 thermocycler, with an initial denaturation of 5 min at 94ºC, followed by 40 cycles of: 30 s at 94ºC; 30 s at 36ºC and 1 min at 72ºC, and a final extension of 3 min at 72ºC. Following amplification, the PCR products were electrophoresed in a 1.4% agarose gel. The gel was stained with ethidium bromide and photographed under UV light.
Data analysis
RAPD bands were used to generate a binary (presence/absence) matrix, from which a genetic similarity matrix was computed using the Jaccard coefficient, as implemented in NTSYS 2.11 (Numerical Taxonomy System of Multivariate Programs). A significance test between the genetic similarity matrix and the cophenetic matrix obtained from the dendrogram was carried out using the Mantel test (25). A Principal Coordinates Analysis (PCO) was conducted based on the genetic similarity matrix to assess the distribution and clustering of isolates. The UPGMA (Unweighted Pair-Group Method with Arithmetical Average) hierarchical method was used to group the units and to build the dendrogram. The software Bood 3.04 (6) was used for bootstrap analysis, which asses the reliability of each grouping after 10.000 pseudo replicates.
RESULTS AND DISCUSSION
A total of 157 colonies were isolated from salivary samples of the 40 studied individuals, using the colony morphology in MSBS as an indicator. Sixty-four of those isolates were characterized biochemically as S. mutans and 10 as S. sobrinus. All analyzed individuals showed evidence of the presence of S. mutans in their salivary samples, with the number of isolates of S. mutans per individual varying among one (20 individuals), two (16 individuals), and three (4 individuals). Ten isolates of S. sobrinus were obtained from individuals that had been colonized by both species.
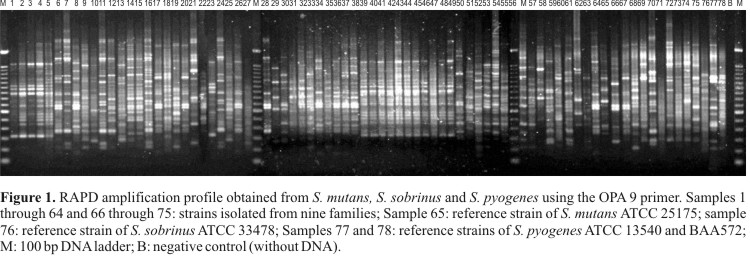
Five primers were used to characterize these isolates for a total of 101 reliable bands. The presence of genetic polymorphism was observed based on the amplification profiles (Fig. 1). The analysis of these profiles generated a dendrogram that separated the isolates into only two groups of high genetic similarity based on reliable bootstrap values (Fig. 2).
Group I included 11 isolates of S. mutans from family 6 (SM38 through SM48), with 75% genetic similarity and 99% bootstrap support, as well as several isolates with 100% genetic identity (SM38 and SM40, SM45 and SM46). Isolates SM1 and SM2, with 100% genetic similarity, were obtained from the same individual. Some isolates were grouped into small clusters of low similarity but high bootstrap values (above 90%): SM3 and SM4; SM9 and SM10; SM35, SM36 and SM37; SM32 and SM34; SM51 and SM52.
The 74 strains isolated in the present study showed high variability when comparing with reference strains (ATCC 25175 of S. mutans and ATCC 33478 of S. sobrinus). There was no concordance between the molecular and biochemical characterization of the analyzed oral streptococci, given that the RAPD markers only grouped the SS75 strain with the reference strain of S. sobrinus.
Interestingly, Truong et al. (38) analyzed reference strains of oral streptococci using RAPD for the identification and sub-classification of all the studied strains and suggested that RAPD can be valuable to discriminate the species S. mutans and S. sobrinus, both from each other and from other oral streptococci. However, similarly to those authors, the present study indicated that the RAPD markers discriminated the strains ATCC 13540 and BAA572 of S. pyogenes, forming a separate group (Group II, Fig. 2).
The principal coordinate analysis (Fig. 3) grouped together isolates from families 6 (I), 5 (IV), and 1 (III), explaining 16% of the total variance in the dataset. Therefore, the large diversity among salivary samples of S. mutans and S. sobrinus from the individuals of the nine analyzed families justified the different forms and intensities of the clinical manifestation of caries in this population. Of the 74 isolates, 36 belonged to individuals that showed active caries at the time of collection, yet these isolates did not show a consistent mutual similarity based on the cluster analysis.
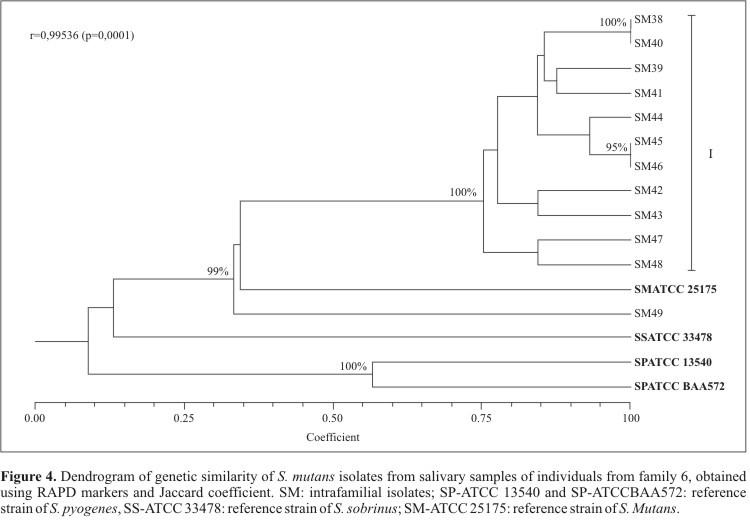
On the other hand, when analyzing isolates from family 6 alone, group I showed approximately 75% similarity (100% bootstrap support) (Fig. 4). In this cluster, isolates SM38 and SM40 (obtained from the mother and the father, respectively) showed 100% genetic similarity (100% bootstrap support). A similar pattern was observed in the case of isolates SM45 and SM46, which were obtained from two brothers of the same family (95% bootstrap support). These observations are consistent with the occurrence of intra-familial transmission. In that family, only two individuals (sons 3 and 4) showed active caries and shared isolates with high genetic similarity, possibly indicating the existence of a more virulent biosorotype of S. mutans. Moreover, son 3 reported a high consumption level of saccharose, whereas son 4 had a low frequency of dental brushing, two important causal components to determine the occurrence of this characteristically multifactorial disease.
The reference strain of S. mutans, SM-ATCC 25175, showed a 33% genetic similarity with these isolates (99% bootstrap support, Fig. 4). However, one of the isolates (SM49) showed a particularly large genetic distance in relation to the other isolates. The reference strain of S. sobrinus, SS- ATCC 33478, showed a genetic dissimilarity level of 87% in relation to the S. mutans isolates. Also, isolates SM45 and SM46, in spite of showing 100% genetic similarity, presented distinct colonial morphologies, demonstrating that morphological differences among isolates are not necessarily indicative of underlying genetic differences. However, this pattern needs to be further studied given the low number of isolated colonies per individual in the present study. Several authors (9,24,30) reported that, in the case of highly selective media, only the bacteria with high concentration in the saliva are often isolated, and therefore other isolates might also be present but undetected.
The intra-familial distribution of this cariogenic microorganism was different among families, a pattern already reported by Emanuelsson et al. (10). These results are consistent with those of Spolidorio et al. (37), who suggested that members of the same family can harbor different bacterial genotypes. The present study also indicates that multiple infections might have occurred in the same individuals, as has been suggested by the high genetic variability detected using RAPD markers.
In conclusion, the high genetic variability among isolates of S. mutans within and among families and with different patterns of intra-familial distribution suggested the occurrence of multiple infections. Moreover, the RAPD analysis suggests that the biochemical marker might not be efficient in the identification of S. mutans and S. sobrinus. Finally, there was no relationship between the morphological and biological characteristics of the isolates and their genetic proximity based on the RAPD markers. The data indicated intra-familial transmission due to the presence of isolates with 100% genetic similarity in different members of the same families.
Submitted: March 18, 2007; Returned to authors for corrections: May 20, 2007; Approved: November 15, 2007.
- 1. Bowden, G.H.; Hamilton, I.R. (1998). Survival of oral bacteria. Crit. Rev. Oral Biol. Med., 9, 54-85.
- 2. Bratthall, D. (1992). Caries, views and perspectivies. Scand. J. Dent. Res., 100, 47-51.
- 3. Caufield, P.W.; Cutter, G.R.; Dasanayake, A.P. (1993). Initial acquisition of mutans streptococci by infants: Evidence for a dscrete window of infectivity. J. Dent. Res., 72, 37-45.
- 4. Caufield, P.W.; Ratanapridakul, K.; Allen, D.N.; Cutter, G.R. (1988). Plasmid-containing strains of Streptococcus mutans cluster within family and racial cohorts: implications for natural transmission. Infect. Immun., 56, 3216-3220.
- 5. Caufield, P.W.; Walker, T.M. (1989). Genetic diversity within Streptococcus mutans evident from chromosomal DNA restriction fragment polymorphisms. J. Clin. Microbiol., 27, 274-278.
- 6. Coelho, A.S.G. (2005). Avaliação de dendrogramas baseados em estimativas de distâncias/similaridades genéticas através do procedimento de bootstraps BOOD 3.03. Departamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Goiás.
- 7. Davey, A.L.; Rogers, A.H. (1984). Multiple types of the bacterium Streptococcus mutans in the human mouth and their intrafamily transmission. Arch. Oral Biol., 29, 453-460.
- 8. De Hoog, G.S.; Attili-Angelis, D.; Vicente, V.A.; Queiroz-Telles, F. (2004). Molecular ecology and pathogenic potencial of Fonsecaea species. Medical Mycology., 42, 405-416.
- 9. Duchin, S.; Van Houte, J. (1978). Colonization of teeth in human by Streptococcus mutans as related to its concentration in saliva and host age. Infect. Immun., 20, 120-125.
- 10. Emanuelsson, I.R.; Li, Y.; Bratthall, D. (1998). Genotyping shows different strains of mutans streptococci between father and child and within parental pairs in Swedish families. Oral Microbiol. Immunol, 13, 271-277.
- 11. Gilmour, M.N.; Whittam, T.S.; Kilian, M.; Selander, R.K. (1987). Genetic relationships among the oral streptococci. J. Bacteriol., 69, 5247-5257.
- 12. Gold, O.G.; Jordan, H.V.; Van Houte, J. (1973). A selective medium for Streptococcus mutans. Arch. Oral Biol., 18, 1357-1364.
- 13. Gonçalves, R.B.; Rosa, R.T.; Stipp, R.N.; Napimoga, M.H.; Rosa, E.A.R.; Höfling, J.F. (2002). Genetic diversity and cariogenic potential of Streptococcus mutans. J. Dent. Res., 81, 281-286.
- 14. Grönroos, L.; Alaluusua, S. (2000). Site-specific oral colonization of mutans streptococci detected by arbitrarily primed PCR fingerprinting. Caries Res., 34, 474-80.
- 15. Huang, X.; Liu, T.; Cheng, G. (2001). Typing of Streptococcus mutans (serotype C) by arbitrarily primed polymerase chain reaction. Zhonghua Kou Qiang Yi Xue Za Zhi. 36(4), 281-284.
- 16. Igarashi, T.; Ichikawa, K.; Yamamoto, A.; Goto, N. (2001). Identification of mutans streptococcal species by the PCR products of the dex genes. J. Microbiol. Methods., 46(2), 99-105.
- 17. Jordan, H.V.; Van Hote, R.J. (1985). Transmission of the oral bacterium Streptococcus mutans to young Macaca fascicularis monkeys from human nursey attendants. Arch. Oral Biol., 30, 863-864.
- 18. Kamiya, R.U. (2003). Análise genotípica e mutacinotipagem de Streptococcus mutans isolados de indivíduos cárie-ativos e livres de cárie Thesis, Piracicaba, São Paulo: UNICAMP.
- 19. Klein, M.I.; Florio, F.M.; Pereira, A.C.; Höfling, J.F.; Gonçalves, R.B. (2004). Longitudinal study of transmission, diversity and stability of Streptococcus mutans and Streptococcus sobrinus genotypes in Brazilian nursery children. J. Clin. Microbiol., 42, 4620-4626.
- 20. Koga, C.Y.; Unterkircher, C. S. Fantinato, V.; Shimizu, M.T.; Jorge, A.O.C. (1995). Testes de atividade de cárie. Avaliação de diferentes métodos. RGO, 43(3), 141-44.
- 21. Li, Y.H.; Lau, P.C.Y.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. (2001). Natural genetic transformation of Streptococus mutans growing in biofilms. J. Bacteriol, 183, 897-908.
- 22. Li, Y.; Caufield, P.W. (1995). The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J. Dent. Res., 74, 681-685.
- 23. Li, Y.; Wang, W.; Caufield, P.W. (2000). The fidelity of mutans streptococci transmission and caries status correlate with breast-feeding experience among Chinese families. Caries Res., 34, 123-132.
- 24. Lindquist, B. (1991). Mutans streptococci in human dentition Thesis, Göteborg: University of Göteborg.
- 25. Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res., 27, 209-220.
- 26. Marchant, S.; Brailsford, S.R.; Twomey, A.C.; Roberts, G.J.; Beighton, D.I. (2001). The predominant microflora of nursing caries lesions. Caries Res., 35(6), 397-406.
- 27. Masuda, N.; Shimamoto, T.; Kitamura, K.; Sobue, S.; Hamada, S. (1985). Transmission of Streptococcus mutans in some selected families. Microbios, 44, 223-232.
-
28Ministério da Saúde, Saúde Bucal, Programa Brasil Sorridente, Departamento de Atenção Básica. Available at: http://www.saude.gov.br/dab/saudebucal/
» link - 29. Napimoga, M.H.; Kamiya, R.U.; Rosa, R.T.; Rosa, A.R.; Höfling, J.F.; Mattos-Graner, R.O.; Gonçalves, R.B. (2004). Genotypic diversity and virulence traits of Streptococcus mutans in caries-free and caries-active individuals. Journal of Medical Microbiology, 53, 697-703.
- 30. Newbrun, E. (1988). Cariologia Santos, São Paulo.
- 31. Oho, T.; Yamashita, Y.; Shimazaki, Y.; Kushiyama, M.; Koga, T. (2000). Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol., 15(4), 258-62.
- 32. Okada, M.; Soda, Y.; Hayashi, F.; Doi, T.; Suzuki, J.; Miura, K.; Kozai, K. (2002). PCR detection of Streptococcus mutans and S. sobrinus in dental plaque samples from Japanese pre-school children. J. Med. Microbiol., 51, 443-447.
- 33. Pereira, M.S.; Leal, N.C.; Sobreira, M.; Almeida, A.M.P.; Siqueira-Júnior, J.P.; Takak, G.M. (2002). Typing human bovine Sthaphylococcus aureus by RAPD-PCR and ribotyping-PCR. Letters in Applied Microbiology, 35, 32-36.
- 34. Redmo-Emanuelsson, I.M.; Thornqvist, E. (2001). Distribution of mutans streptococci in families: a longitudinal study. Acta Odontol. Scand., 59, 93-98.
- 35. Redmo-Emanuelsson, I.M.; Wang, X.M. (1998). Demonstration of identical strains of mutans streptococci within Chinese families by genotyping. Eur. J. Oral Sci., 106, 788-794.
- 36. Saarela, M.; Von Troil-Lindén, B.; Torkko, H.; Stucki, A.M.; Alaluusua, S.; Jousimies-Somer, H.; Asikainen, S. (1993). Transmission of oral bacteria species between spouses. Oral Microbiol. Immunol., 8, 349-354.
- 37. Spolidorio, D.M.P.; Höfling, J.F.; Pizzolitto, A.C.; Rosa, E.A.; Negrini, T.C.; Spolidorio, L.C. (2003). Genetic polymorphism of Streptococcus mutans in brazilian family members. Braz. J. Microbiol, 34(3), 213-217.
- 38. Truong, T.L.; Ménard, C.; Mouton, C.; Trahan, L. (2000). Identification of mutans and other oral streptococci by random amplified polymorphic DNA analysis. J. Med. Microbiol, 49, 63-71.
Publication Dates
-
Publication in this collection
28 Jan 2008 -
Date of issue
Dec 2007
History
-
Accepted
15 Nov 2007 -
Reviewed
20 May 2007 -
Received
18 Mar 2007