Abstracts
Dermatophytes, capable to use keratin of the host for nutrition, belong to one of the major groups of pathogenic fungi. Since dermatophytes are a closely related group they share various common features, and the morphology of isolates of a given species can be atypical, making species identification and differentiation even more difficult. Many methods have been explored in attempts to distinguish dermatophytes, but the combined use of different approaches for the investigation of the intraspecific and interspecific variability of Trichophyton continues to be scarce. Some studies have shown that amplified fragments of the small ribosomal DNA subunit 18S contains variable regions which can be used to discriminate between medically relevant yeast species, indicating that these regions could also be used for differentiation between dermatophytes. In our study, sequence analysis of the 18S-rDNA gene was combined with morphological and biochemical criteria in order to detect genetic differences between seven Trichophyton isolates and estimate their phylogenetic relationships. The results show that the isolates investigated belong to the Trichophyton group, which potentially contains the Trichophyton rubrum cluster.
Trichophyton; 18S-rDNA sequencing; extracellular enzymes; dermatophyte
Os dermatófitos formam um dos principais grupos de fungos patogênicos, caracterizados pela utilização da queratina do hospedeiro para sua nutrição. Por se constituírem um grupo de fungos intimamente relacionados, compartilham uma série de características comuns. Além disto, a morfologia de isolados de determinadas espécies pode ser atípica, tornando a identificação das espécies ainda mais difícil. Muitos métodos vêm sendo explorados na tentativa de distinguir dermatófitos, porém a associação de diferentes abordagens para a investigação da variabilidade intra e interespecífica de Trichophyton permanece escassa. Alguns trabalhos têm demonstrado que apesar de conservados, os fragmentos amplificados da seqüência correspondente à subunidade ribossomal menor 18S, contêm regiões conhecidas por sua variabilidade e capacidade de distinção entre espécies de levedura de importância médica, indicando que esta região também pode ser útil na diferenciação dos dermatófitos. Nesse estudo, a análise da seqüência do DNA ribossomal 18S foi combinada com critérios morfológicos e bioquímicos com o objetivo de se detectar possíveis diferenças genéticas entre sete isolados e estimar suas relações filogenéticas. Os resultados mostram que os isolados investigados pertencem ao grupo Trichophyton, o qual pode potencialmente conter o cluster Trichophyton rubrum.
Trichophyton; seqüenciamento do DNA ribossomal 18S; enzimas extracelulares; dermatófito
18S-rDNA SEQUENCING, ENZYME PATTERNS AND MORPHOLOGICAL CHARACTERIZATION OF TRICHOPHYTON ISOLATES
Adriana Mendes do Nascimento; Nilce M. Martinez-Rossi* * Corresponding author. Mailing address: Departamento de Genética, Faculdade de Medicina de Ribeirão Preto, USP. Av. Bandeirantes, 3900. 14049-900, Ribeirão Preto, SP, Brasil. Fax: (+5516) 633-0069. E-mail: nmmrossi@fmrp.usp.br
Departamento de Genética, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brasil
Submitted: March 09, 2001; Returned to authors for corrections: May 17, 2001; Approved: July 23, 2001
ABSTRACT
Dermatophytes, capable to use keratin of the host for nutrition, belong to one of the major groups of pathogenic fungi. Since dermatophytes are a closely related group they share various common features, and the morphology of isolates of a given species can be atypical, making species identification and differentiation even more difficult. Many methods have been explored in attempts to distinguish dermatophytes, but the combined use of different approaches for the investigation of the intraspecific and interspecific variability of Trichophyton continues to be scarce. Some studies have shown that amplified fragments of the small ribosomal DNA subunit 18S contains variable regions which can be used to discriminate between medically relevant yeast species, indicating that these regions could also be used for differentiation between dermatophytes. In our study, sequence analysis of the 18S-rDNA gene was combined with morphological and biochemical criteria in order to detect genetic differences between seven Trichophyton isolates and estimate their phylogenetic relationships. The results show that the isolates investigated belong to the Trichophyton group, which potentially contains the Trichophyton rubrum cluster.
Key words:Trichophyton, 18S-rDNA sequencing, extracellular enzymes, dermatophyte
INTRODUCTION
With the significant increase in the incidence of mycoses, especially in immunosuppressed patients, information about the variability of dermatophyte species and the evolutionary relationship between them is becoming increasingly more important for the understanding of the mechanisms of pathogenicity. Such knowledge is also fundamental for the refinement of techniques for the detection and control of pathogens (7).
Although morphological appearance is essential for fungal taxonomy in general, it alone is insufficient for species identification or differentiation (3) because in addition to sharing a series of morphological characteristics, some species may also present atypical variations resulting from sensitivity to culture conditions. Thus, a combination of morphology and genetic and pathogenicity testing has been used to establish the taxonomy of fungal groups such as Alternaria (1) and Metarhizium (9), and secondary metabolites patterns have been of help in differentiating between Penicillium species (15).
DNA sequence data can also be extremely useful for differentiating between fungal species. Molecular analyses of ribosomal DNA sequences has permitted inferences to be made on the relationship between different levels of divergence and is being increasingly explored as a taxonomic tool (4). Analysis of ribosomal nucleotide sequences is well suited for use as taxonomic tool because they are universally present and their function in protein synthesis is highly conserved both in prokaryotes and eukaryotes (31).
Despite their medical importance, few studies are available on the intraspecific and interspecific variability of Trichophyton and other dermatophytes and for the clarification of aspects of their phylogeny. The present study investigated the enzymatic patterns, morphological features and nucleotide sequences of 18S-rDNA in 7 Trichophyton isolates. The results obtained were pooled and analyzed from a phylogenetic viewpoint.
MATERIALS AND METHODS
Fungal isolates and growth conditions
The clinical Trichophyton isolates used in the present study were denoted as H2, H4, H6 (14,24) 58, 07, 75 and 110, and were obtained from patients admitted to the University Hospital of Ribeirão Preto, São Paulo state, Brazil. For microscopic examination the isolates were cultured on Sabouraud-agar for 5 days at 28ºC by the slide culture method (26). For DNA extraction the isolates were cultured in Sabouraud broth at 28ºC for 72 hours. Punch biopsies (10 mm diameter) made from thalli grown in minimum medium (12) supplemented with 1% casamino acid solution were used as inoculum. Sabouraud broth was also used to evaluate enzyme production of the different Trichophyton isolates, duplicate cultures being incubated at 28ºC for 2 or 4 weeks.
Enzymatic extract and quantification
The aqueous phase of the broth culture was centrifuged at 3x103 g for 10 min at 4ºC and the level of extracellular alkaline phosphatase activity in the supernatant (free from mycelia and conidia) detected using polyacrylamide gel electrophoresis (PAGE). Other enzymes were detected with the Api-Zym® enzymatic quantification test (Api system, Montalieu Vercieu, France), using 50 µL of broth in each well incubated at 37ºC for 4 hours. The Api-Zym® is a semi-quantitative micromethod designed for the research of enzymatic activities applicable to all specimens, including tissues, cells, biological fluids, microorganisms, etc (details at: http://biomerieux.com). The Api-Zym® test can detect 19 different enzymes and score their concentrations on a rating scale of 0-5, scoring being done using the Api-Zym® colour scale, on which 0 = no enzyme, 1 = 5 nmol, 2 = 10 nmol, 3 = 20 nmol, 4 = 30 nmol and 5 = 40 nmol or more. To detect intracellular alkaline phosphatase activity about 1 g of dry mycelium (from filtration of the broth culture) was macerated in 10 mL of 0.3 M Tris-HCl extration buffer (pH 8.0) and the homogenate centrifuged at 2 x 104g for 10 min at 4ºC. The activity of the released intracellular alkaline phosphatase in the supernatant was detected on a polyacrylamide gel gradient.
Polyacrylamide Gel Gradient Electrophoresis
Aliquots of 35 µL of centrifuged broth culture (containing about 3 enzyme mU) were mixed with 15 µL of diluting buffer (0.5 M Tris, pH 6.7, 30% glycerol, and bromophenol blue) and applied to a polyacrylamide gel gradient (6-15%). Electrophoresis was performed with a continuous current of 25 mA at 4ºC for 9 hours in Tris-glycine buffer [2.8% Tris-HCl (m/v) and 0.6% glycine buffer (m/v), pH 8.3]. After electrophoresis, the gels were submerged in 0.1M Tris buffer, pH 8.4, containing 5 mM a-naphthylsulfate and incubated for about 30 min at 37ºC, the reaction being stopped by the addition of 10% trichloroacetic acid (18).
PCR amplification and sequencing
The genomic DNA was extracted by the method of Andrade-Monteiro et al. (2). The 18S rDNA sequences were amplified by PCR using the NS1a and NS4 primers designed using 18S ribosomal gene sequences obtained from the GenBank data bank, whose accession numbers are given in Table 1. Primers NS1a (5'-GGTCTTGTAATTGGAATGAG-3') and NS4 (5'-CTTCCGTCAATTCCTTTAAG-3') were synthesized by DNAgency, Pennsylvania, USA. Amplification was performed in a 20 µL volume containing 10 mM Tris-HCl (pH 9.0), 0.2 mM of each dNTP (Pharmacia Biotech, Uppsala, Sweden), 2 pmol of each primer, 0.5 U of AmpliTaq® DNA polymerase (Perkin-Elmer, CT, USA), and about 10 ng DNA. PCR amplification was carried out under the following conditions: an initial incubation of 5 min at 94ºC followed by 3 cycles of 20 s at 94ºC (denaturation), 20 s at 59ºC (annealing) and 50 s at 72ºC (extension) followed by 30 cycles of 20 s at 94ºC, 20 s at 56ºC and 50 s at 72ºC. The amplification products were resolved by electrophoresis on 1% low-melting point agarose gel (United States Biochemical, Ohio, USA) in TA buffer (0.1 M Tris-HCl and 12.5 mM NaAc, pH 8.0) to verify the fragment sizes, before being used as template in reamplification reactions. The reamplification reactions were made by transferring 2µL of the original reaction to a fresh PCR reaction mix and reamplified under the same PCR conditions as above. Bands corresponding to the fragments of interest (~700 bp) were cut out from the agarose gel and the DNA was recovered by passive elution (27) for subsequent use in sequencing reactions. Sequencing of the 18S rDNA region was performed by the enzymatic method of Sanger et al. (28) on double-stranded molecules using the CircumVent Thermal Cycle DideoxyDNA Sequencing kit (New England - BioLabs) and dATP radioactively labeled with a [35S] for 25 cycles of 20 s at 94ºC, 20 s at 48ºC and 40s at 70ºC, two sequencing reactions being performed for each strain from two different PCR products. The DNA fragments produced by the sequencing reactions were separated by electrophoresis on a 0.4-mm thick denaturing gel consisting of 5% (m/v) LongRanger polyacrylamide (FMB - Bioproducts) and 7 M urea in 1.2 X TBE buffer (89 mM Tris-borate and 0.2 mM EDTA, pH 8.0) for 5 hours (short run) and 10 hours (long run) at 55 Watts.
Sequence Analysis
The sequences were aligned using as reference the 18S-rDNA sequence of T. rubrum (6). The character matrix consisted of the 18S rDNA sequences of 13 isolates of dermatophyte fungi, with the sequences of the species Epidermophyton floccosum and Blastomyces dermatiditis being used as external groups. The resulting matrix was analyzed by the parsimony technique using the branch-and-bound option of the PAUP software (30) on the basis of 604 characters and 8 phylogenetically informative positions. The events of character transformation were estimated using the MacClade software (19).
RESULTS
Morphology
All isolates investigated had white colonies that varied in consistency and texture, with isolates H2, H6, 58, 75 and 07 having a cotton-like appearance and isolates H4 and 110 a velvety appearance. Pigmentation on the reverse side was practically imperceptible in the young colonies, but on about the 9th day of culture isolates H2, H4, 58 and 75 started to show a reddish color on the base of their colonies, which did not occur with isolates 110, H6 or 07. Microscopic examination of all isolates revealed the morphological pattern expected for the species Trichophyton rubrum (11,17).
Enzyme Polymorphism
Fig. 1A shows the intracellular, and Fig. 1B the extracellular, electrophoretic patterns of the enzyme alkaline phosphatase from the different isolates cultured in Sabouraud broth for 2 weeks. Only 1 band was detected for this enzyme in all isolates, although the electrophoretic mobility of the alkaline phosphatase produced and secreted by isolate 07 differed from that of the other isolates, producing 2 distinct isoenzyme patterns.
Enzyme quantification
The profiles and quantification of extracellular enzymes obtained from Trichophyton isolates are shown in Table 2. Alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, b-glucosidase, N-acetyl-b-glucosaminidase, and a-mannosidase were present in cultures of all isolates. Under the culture conditions used, no isolate produced detectable levels of trypsin, chymotrypsin, a-galactosidase, b-galactosidase, b-glucuronidase or a-fucosidase. Valine arylamidase and cystine arylamidase were present only in cultures of isolate 58 at an estimated concentration of 5 to 10 nmol. Lipase (C14) was detected in isolate H6, also at about 5 to 10 nmol, and a-glucosidase was detected in isolate 07 at about 5 nmol.
Alkaline phosphatase was the enzyme most abundantly released by all isolates at a level of about 40 nmol. Quantitative differences in enzyme release occurred between the isolates tested, with scores ranging from 0 to 5 for b-glucosidase and a-mannosidase, from 1 to 5 for leucine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase, from 1 to 4 for esterase lipase (C8), from 2 to 3 for esterase (C4), and from 2 to 5 for N-acetyl- b-glucosaminidase.
For most isolates the release of esterase (C4), esterase lipase (C8), acid phosphatase, b-glucosidase and a-mannosidase appeared to increase as a function of culture time, in contrast to leucine arylamidase and N-acetyl- b-glucosaminidase, whose activities or release appeared to decrease in some isolates after prolonged growth.
Phylogenetic Analysis
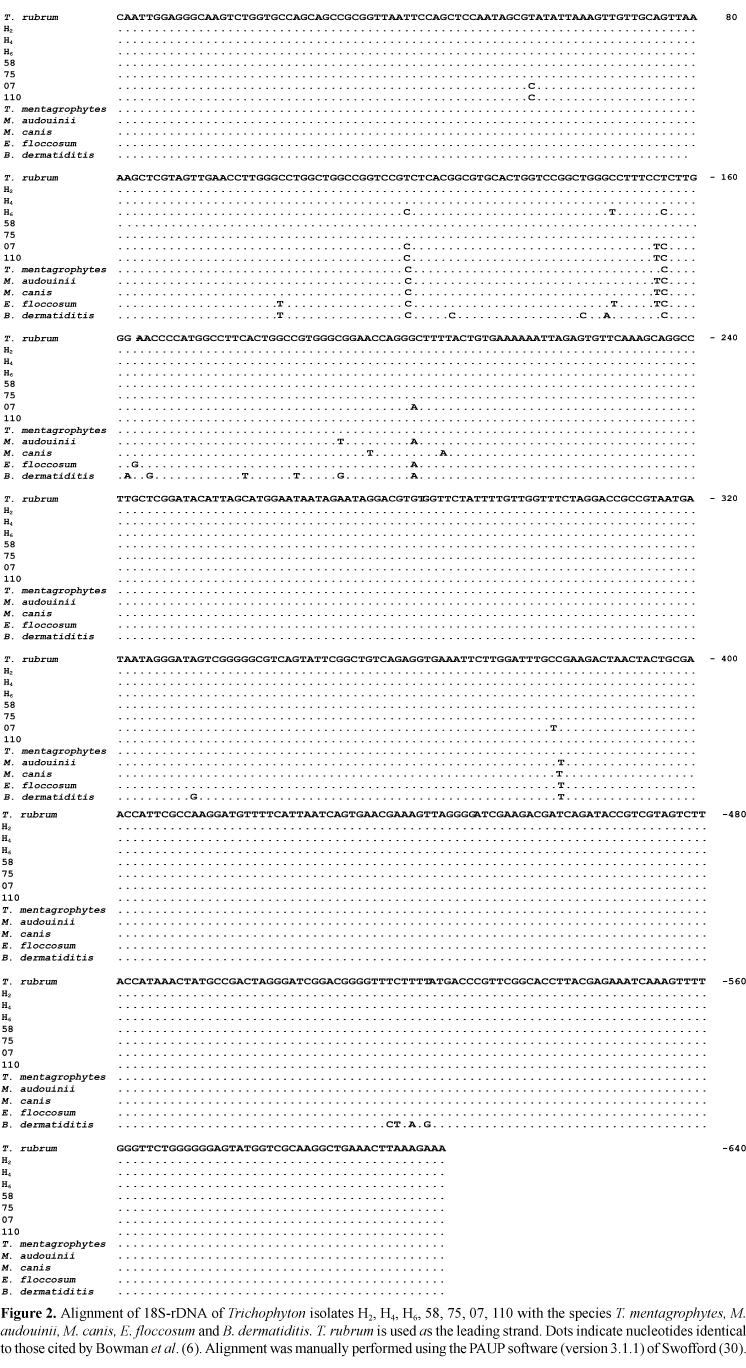
Partial sequences of approximately 604 bp, corresponding to the smaller subunit of gene 18S rDNA of each isolate, were obtained and aligned with sequences deposited in the GenBank data bank using T. rubrum (6) as an alignment model (Fig. 2). A high degree of homology was observed between the sequences described in the literature and those determined in the present study. Table 3 shows the distance between taxa according to the absolute number of different bases (absolute distances) and the percentage of divergent bases in relation to the total number of bases sequenced (mean distances).
A dendrogram (Fig. 3) was obtained based on the maximum parsimony principle using the branch-and-bound option of the PAUP 3.1.1 software as the method to estimate the phylogenetic relationship between taxa. The most parsimonious tree consisted of 13 taxa and presented a length of 23 (on the basis of 8 phylogenetically informative positions), a consistency index of 0.913, and a homoplasy index of 0.087.
The partial sequences corresponding to the smaller subunit of gene 18S rDNA of isolates H2, H4, H6, 58, 07, 75 and 110 have been deposited in GenBank under the accession numbers AF338187, AF338188, AF338189, AF338185, AF338183, AF338186, AF338184, respectively.
DISCUSSION
Few studies are available about the intraspecific and interspecific variability of Trichophyton and other dermatophytes or on the evolutionary relationship between them. This information is essential for the development and refinement of systems for pathogen identification using DNA probes or amplification with species-specific primers. Primers which are specific for fungi and do not hybridize to DNA of other eukaryotes or prokaryotes have been developed and allow specific amplification of fungal DNA from human tissue samples containing fungi (5, 13, 21, 25).
It has been reported that, despite a high degree of conservation, amplified fragments of the 18S-rDNA gene contain regions which are known for their variability and discriminating capacity within medically relevant yeast species (20,22) and there seem to be differences in the 18S-rRNA sequences even within the species Cryptococcus neoformans (5,10). This evidence supports the potential use of these ribosomal DNA regions for differentiation between dermatophytes.
However, the amplified 18S-rDNA sequences proved to be quite conserved among the different isolates analyzed, offering a relatively low resolution of the Trichophyton group. The sequences of isolates H2, H4, 58 and 75 were indistinguishable from the sequence of the homologous region of T. rubrum (6), indicating that this region is highly conserved in these organisms and that the latter probably belong to a phylogenetically homogeneous group. The sequence of isolate H6 was very similar to that of T. mentagrophytes, and differed in only 3 nucleotides from the homologous sequence of T. rubrum described by Bowman et al. (6). Isolate H6 produced lipase like T. mentagrophytes, but production was low and there is no evidence that the ability to produce this enzyme is an exclusive characteristic of T. mentagrophytes, so it is not possible to state that isolate H6 is T. mentagrophytes.
Isolates 110 and 07 proved to be more closely related to each other than the remaining isolates belonging to the genus Trichophyton. Although isolate 07 was also similar to the species M. canis and M. audouinii, it cannot be excluded from the Trichophyton group since it was assigned to one of the terminal branches of this group (based on 18S sequence analysis).
Lipase secretion (C14) detected only in isolate H6, secretion of valine arylamidase and cystine arylamidase by isolate 58 and the alkaline phosphatase polymorphism (Fig. 1A and B) presented by isolate 07 are features which indicate the variability occurring among these dermatophytes and may represent properties that permit the differentiation between species and subspecies.
The isolates investigated were also able to secrete a variety of extracellular enzymes with different patterns. The secretion of N-acetyl-b-glucosaminidase and alkaline phosphatase, detected at high levels in all isolates, seems to be essential for the growth of these fungi. These enzymes may also be involved in the process of tinea development since they can metabolize a series of substrates including different human skin components. Similar results were obtained for T. rubrum (8), M. canis (23) and Candida albicans (29).
As was the case with morphological criteria, the exclusive use of biochemical characteristics was not sufficient to differentiate between isolates. However, when combined, the enzymatic patterns and the results of 18S sequences analysis were coherent, showing that the isolates investigated probably belong to the Trichophyton group, which seems to contain the T. rubrum subgroup formed by exclusion of isolates 07, 110 and H6. This relationship, illustrated in Fig. 3, is supported by morphological evidence since the isolates belonging to the supposed T. rubrum group exhibited red pigmentation on the base of the colonies while the remaining isolates exhibited a yellowish, or no pigmentation. However, the lack of red pigmentation can also be observed in some isolates of this species, especially those obtained from patients who had been treated with griseofulvin for some time (17).
Morphological examination revealed variations, especially with respect to colony texture, but the general macroscopic appearance and microscopic morphology of the isolates investigated were consistent with the patterns reported in the literature for T. rubrum, the variations detected may have been the result of random mutations in these isolates. Gräser et al. (16) reported that 96 T. rubrum strains displaying different colony morphologies did not reveal any DNA polymorphism when analyzed using molecular markers, suggesting a strictly clonal mode of reproduction and a strong adaptation to human skin.
The present results show that the Trichophyton group is heterogeneous and that new isolates and other genome regions need to be analyzed to elucidate the taxonomic relationships of a group that shows differences in phenotype but are very similar in their ecology.
ACKNOWLEDGMENTS
This work was supported by grants from FAPESP, CNPq and FAEPA. We are deeply grateful to Prof. A.R. Templeton (Washington University, St Louis, MO, USA) for his warm hospitality during A. M. N.'s stay in his laboratory, where part of this work was carried out. We also thank Dr. R.O.A.A. Brito for research assistance and Mrs. Elettra Greene for revising the manuscript.
RESUMO
Seqüenciamento do DNA ribossomal 18S, padrões enzimáticos e caracterização morfológica de isolados de Trichophyton
Os dermatófitos formam um dos principais grupos de fungos patogênicos, caracterizados pela utilização da queratina do hospedeiro para sua nutrição. Por se constituírem um grupo de fungos intimamente relacionados, compartilham uma série de características comuns. Além disto, a morfologia de isolados de determinadas espécies pode ser atípica, tornando a identificação das espécies ainda mais difícil. Muitos métodos vêm sendo explorados na tentativa de distinguir dermatófitos, porém a associação de diferentes abordagens para a investigação da variabilidade intra e interespecífica de Trichophyton permanece escassa. Alguns trabalhos têm demonstrado que apesar de conservados, os fragmentos amplificados da seqüência correspondente à subunidade ribossomal menor 18S, contêm regiões conhecidas por sua variabilidade e capacidade de distinção entre espécies de levedura de importância médica, indicando que esta região também pode ser útil na diferenciação dos dermatófitos. Nesse estudo, a análise da seqüência do DNA ribossomal 18S foi combinada com critérios morfológicos e bioquímicos com o objetivo de se detectar possíveis diferenças genéticas entre sete isolados e estimar suas relações filogenéticas. Os resultados mostram que os isolados investigados pertencem ao grupo Trichophyton, o qual pode potencialmente conter o cluster Trichophyton rubrum.
Palavras-chave:Trichophyton, seqüenciamento do DNA ribossomal 18S, enzimas extracelulares, dermatófito.
- 1. Andersen, B.; Thrane, U. Differentiation of Alternaria infectoria and Alternaria alternata based on morphology, metabolite profiles, and cultural characteristics. Can. J. Microbiol., 42:685-689, 1996.
- 2. Andrade-Monteiro, C.; Maccheroni Jr., W.; Rossi, A.; Martinez-Rossi, N.M. A simplified method for the isolation of high molecular weight DNA from Aspergillus nidulans. Brazil. J. Genetics, 17:447-448, 1994.
- 3. Attili D.S.; De Hoog, G.S.; Pizzirani-Kleiner, A.A. rDNA-RFLP and ITSI sequencing of species of the genus Fonsecaea, agents of chromoblastomycosis. Medical Mycology, 36:219-225, 1998.
- 4. Berbee, M.L.; Taylor, J.W. Convergence in Ascospore discharge mechanism among Pyrenomycete fungi based on 18S ribosomal RNA gene sequence. Mol. Phylogenet. Evol., 1:59-71, 1992.
- 5. Bock, M.; Maiwald, M.; Kappe, R.; Nickel, P.; Näher, H. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses, 37:79-84, 1994.
- 6. Bowman, B.H.; Taylor, J.W.; White, T.J. Molecular Evolution of the fungi: Human pathogens. Mol. Biol. Evol, 9:893-904, 1992.
- 7. Bowman, B.H.; White, T.J.; Taylor, J.W. Human pathogenic fungi and their close nonpathogenic relatives. Molec. Phylogenet. Evol., 6:89-96, 1996.
- 8. Brash, J.; Martins, B-S.; Christophers, E. Enzyme release by Trichophyton rubrum dependes on nutricional conditions. Mycoses, 34:365-368, 1991.
- 9. Bridge, P.D.; Williams, M.A.J.; Prior, C.; Paterson, R.R.M. Morphological, biochemical and molecular characteristics of Metarhizium anisopliae and M. flavoviride. J. Gen. Microbiol., 139:1163-1169, 1993.
- 10. Chen, S.; Brownlee, A.; Hunt, C.; Sorrell, T. Strain variation in Cryptococcus neoformans detected as restriction fragment length polymorphisms in amplified ribosomal DNA. XIIth Congress of the International Society for Human and Animal Mycology (ISHAM), Adelaide, 1993, p26.
- 11. Clayton, Y.M.; Midgley, G. Identification of agents of superficial mycoses. In: Evans E.G.V.; Richardson M.D.; (eds). Medical Mycology - a practical approach. Oxford, 1989, p.65-95.
- 12. Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans Biochem. Biophys. Acta, 113:51-56, 1966.
- 13. El Fari, M.L.; Tietz, H.J.; Presber, W.; Sterry, W.; Gräser, Y. Development of an oligonucleotide probe specific for Trichophyton rubrum. British. J. Dermatol., 141:240-245, 1999.
- 14. Fachin, A.L.; Maffei, C.M.L.; Martinez-Rossi, N.M. In vitro susceptibility of Trichophyton rubrum isolates to griseofulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs, Mycopathologia, 135:141-143, 1996.
- 15. Frisvad, J.C.; Filtenborg, O. Terverticillate penicilia: chemotaxononmy and mycotoxin production. Mycologia, 81:836-861, 1989.
- 16. Gräser, Y.; Kühnisch, J.; Presber, W. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum J. Clin. Microbiol., 37:3713-3717, 1999.
- 17. Kwon-Chung, K.J.; Bennett, J.E. Dermatophytoses. In: Lea & Febiger (eds). Medical Mycology. Philadelphia, p.105-161, 1992.
- 18. Maccheroni Jr.; W.; Martinez-Rossi, N.M.; Rossi, A. Does gene palB regulate the transcription or the post-translational modification of Pi-repressible phosphatases of A. nidulans Braz. J. Med. Biol. Res., 28:31-38, 1995.
- 19. Maddison, W.P.; Maddison, D.R. MacClade: interative analysis of phylogeny and character evolution (Sinauer ed). Sunderland, USA, 1992.
- 20. Maiwald, M.; Kappe, R.; Sonntag, H-G. Rapid presumptive identification of medically relevant yeasts to the species level by polymerase chain reaction and restriction enzyme analysis. J. Med. Vet. Mycol., 32:115-122, 1994.
- 21. Makimura, K.; Murayama, S.Y.; Yamaguchi, H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol., 40:358-364, 1994.
- 22. Niesters, H.G.M.; Goessens, W.H.F.; Meis, J.F.M.G.; Quint W.G. Rapid polymerase chain reaction-based identification assays for Candida species. J. Clin. Microbiol., 31:904-910, 1993.
- 23. Papini, R.; Mancianti, F. Extracellular enzymatic activity of Microsporum canis isolates. Mycopathologia, 132:129-132, 1996.
- 24. Pereira, M.; Fachin, A.L.; Martinez-Rossi, N.M. The gene that determines resistance to tioconazole and to acridine derivatives in Aspergillus nidulans may have a corresponding gene in Trichophyton rubrum. Mycopathologia, 143:71-75, 1998.
- 25. Reiss, E.; Tanaka, K.; Bruker, G.; Chazalet, V.; Coleman, D.; Debeaupuis, J.P.; Hanazawa, R.; Latgé, J-P.; Lortholary, J.; Makimura, K.; Morrison, C.J.; Murayama, S.Y.; Naoe, S.; Paris, S.; Sarfati, J.; Shibuya, K.; Sullivan, D.; Uchida, K.; Yamaguchi, H. Molecular diagnosis and epidemiology of fungal infections. Medical Mycology, 36:249-257, 1998.
- 26. Riddell, R.W. Permanent stained mycological preparations obtained by slide culture. Mycologia, 42:265-270, 1950.
- 27. Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A Laboratory Manual New York, Cold Spring Harbor Laboratory, 1989.
- 28. Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA, 74:S463-S467, 1977.
- 29. Sullivan, P.A.; McHugh, N.J.; Romana, L.K.; Shepherd, M.G. The secretion of N-acetylglucosaminidase during germ-tube formation in Candida albicans. J. Gen. Microbiol., 130:2213-2218, 1984.
- 30. Swofford, D.L. PAUP: Phylogenetic analysis using parsimony Version 3.1.1. Smithsonian Institution, Washington, USA, 1993.
- 31. Verweij, P.E.; Meis, J.F.G.M.; Van Den Hurk, P.; Zoll, J.; Samson, R.A.; Melchers, W.J.G. Phylogenetic relationships of five species of Aspergillus and related taxa as deduced by comparison of sequences of small subunit ribosomal RNA. J. Med. Vet. Mycol., 33:185-190, 1995.
Publication Dates
-
Publication in this collection
04 Apr 2002 -
Date of issue
Oct 2001
History
-
Accepted
23 July 2001 -
Reviewed
17 May 2001 -
Received
09 Mar 2001