Abstract
In the Rio Doce basin of southeastern Brazil, the freshwater fish Hoplias malabaricus (trahira) is a widespread predatory characin and one of the few resilient native fishes in a highly impacted lake system. In order to test for genetic differentiation in populations within this basin and for biogeographic relationships among populations of this species in other basins, a study was conducted using RAPD-PCR analysis of Rio Doce samples (N = 63) and phylogeographic analyses with mitochondrial DNA (mtDNA) haplotypes, including the Rio Grande and Macacu river basins. In the Rio Doce basin, the patterns of genetic similarity of RAPD-PCR markers (individual fingerprinting and Nei’s genetic distance) suggest the existence of two genetically different groups, one composed of the lacustrine populations Carioca and Dom Helvécio, and the other of riverine and the remaining lacustrine populations. The differences in the RAPD-PCR patterns may be explained by the existence of sub-basins within this lacustrine system. A maximum parsimony tree of cytochrome b fragment (383 base pairs) supports the view that trahiras of the Rio Doce share a complex biogeographic history with those of neighboring basins. The phylogeographic patterns may be explained by a common history of the watersheds of the Rio Doce, Paraíba do Sul, and Rio Grande basins, corroborating the hypothesis of a Plio-Pleistocene separation of these drainage systems, forming the Mantiqueira "divortium aquarium".
RAPD; SSCP; mtDNA; phylogeography; Hoplias malabaricus; conservation genetics
Phylogeography and RAPD-PCR variation in Hoplias malabaricus (Bloch, 1794) (Pisces, Teleostei) in southeastern Brazil
Jorge A. Dergam1, Samuel R. Paiva2, Carlos E. Schaeffer3, Alexandre L. Godinho4 and Fabio Vieira5
1Departamento de Biologia Animal, Universidade Federal de Viçosa, Viçosa, MG, Brazil.
2Embrapa Genetic Resources and Biotechnology, Brasília, DF, Brazil.
3Departamento de Solos, Universidade Federal de Viçosa, Viçosa, MG, Brazil.
4Departamento de Zoologia - ICB, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil.
5Ecologia, Conservação e Manejo da Vida Silvestre - ICB, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil.
ABSTRACT
In the Rio Doce basin of southeastern Brazil, the freshwater fish Hoplias malabaricus (trahira) is a widespread predatory characin and one of the few resilient native fishes in a highly impacted lake system. In order to test for genetic differentiation in populations within this basin and for biogeographic relationships among populations of this species in other basins, a study was conducted using RAPD-PCR analysis of Rio Doce samples (N = 63) and phylogeographic analyses with mitochondrial DNA (mtDNA) haplotypes, including the Rio Grande and Macacu river basins. In the Rio Doce basin, the patterns of genetic similarity of RAPD-PCR markers (individual fingerprinting and Neis genetic distance) suggest the existence of two genetically different groups, one composed of the lacustrine populations Carioca and Dom Helvécio, and the other of riverine and the remaining lacustrine populations. The differences in the RAPD-PCR patterns may be explained by the existence of sub-basins within this lacustrine system. A maximum parsimony tree of cytochrome b fragment (383 base pairs) supports the view that trahiras of the Rio Doce share a complex biogeographic history with those of neighboring basins. The phylogeographic patterns may be explained by a common history of the watersheds of the Rio Doce, Paraíba do Sul, and Rio Grande basins, corroborating the hypothesis of a Plio-Pleistocene separation of these drainage systems, forming the Mantiqueira divortium aquarium.
Key words: RAPD, SSCP, mtDNA, phylogeography, Hoplias malabaricus, conservation genetics.
Received: February 29, 2002; accepted: July 27, 2002.
INTRODUCTION
The Rio Doce basin is one of the eastern river basins of the State of Minas Gerais, Brazil, with its headwaters located in the Serra do Espinhaço, within the Serra da Mantiqueira mountain system, flowing eastward to the Atlantic Ocean. Also in the western slope of the Serra do Espinhaço lie the headwaters of the Rio Grande basin, which flows westward to form the Paraná river, after its encounter with the Paranaíba river.
Based on morphologic criteria, Godinho and Vieira (1998) identified 77 fish species (37 endemic) in the Rio Doce basin, and 90 species (14 endemic) in the Rio Grande basin. This fish diversity is currently threatened by habitat alterations such as mining, pollution, siltation, and the damming of the main river and its tributaries. Among the diversity-threatening biological factors, the impact of non-indigenous species seems to be the most important one, a situation which applies to the Neotropical region as a whole (Vari and Malabarba, 1998).
The Rio Doce basin is considered a unique river system, due to the occurrence of a complex system of Quaternary lakes in its mid-valley (Petri and Fúlfaro, 1983), formed during the last glaciation maxima, the first one between 14,000 to 10,000 years ago (ya) and thereafter, between 7,800 to 3,360 ya (Tundisi and De Meis, 1985). These lakes have been compared to islands, based on their low levels of similarity in terms of species composition (Sunaga and Verani, 1985). The age of the lacustrine system and the probable existence of different levels of isolation (Figure 1) in the basin, due to barriers such as prominent waterfalls and lakes, make this region a unique scenario for the study of the tempo and mode of evolutionary processes. In this basin, the presence, since 1985, of at least three non-indigenous fish species, the peacock-bass (Cichla ocellaris), an Amazonian cichlid (Astronotus ocellatus), and a piranha species (Pygocentrus piraya), has heavily impacted some of the lacustrine communities (Sunaga and Verani, 1991; Godinho and Formagio, 1992). The absence of juveniles of native fish species in the Dom Helvécio lake was interpreted as evidence of the endangered status of these native populations (Godinho and Formagio, 1992), and the presence of non-indigenous fishes in some lakes was correlated to a 50% reduction in the former richness of species (Godinho et al., 1994). The widespread occurrence of H. malabaricus in the basin and the endangered status of the populations of this species in the lake system led us to perform RAPD-PCR and phylogeographic analyses (Avise et al.,1987; Avise, 1994), as an attempt to understand the role of the lake system in the evolution of the fish communities in the basin, and the historic relationships of the Rio Doce and some of its neighbor basins.
MATERIALS AND METHODS
Fish captures were made in 1993 and 1994, with a total of 63 specimens being gill netted at night or at dusk, in two riverine and four lacustrine sites (Figure 1 and Table I). Lakes Dom Helvécio and Carioca are within the Rio Doce State Park, only the former being currently open for tourism. 12 specimens were collected in the Rio Grande basin in 1994. After capture, a thin slice of muscle tissue was dissected in the field from freshly killed specimens and immediately fixed in 2 mL of absolute ethanol or in 1:1 ethanol: methanol (v:v). DNA extraction followed the CTAB protocol described by Dergam (1996).
Amplifications of RAPD-PCR alleles (Williams et al., 1990) were achieved using the following cycle: 5 min at 95 °C; 1 min at 80 °C; then 45 cycles of 1 min at 92 °C; 1 min at 35 °C; then ramp 37 °C, rising 1 °C every 8 sec, up to 72 °C for 2 min, with a final extension of 7 min at 72 °C. After a pilot test, three primers - A09, A05, and C-13 (Operon Technologies, Inc.) - were selected, based on their consistency in amplifying a large number of scorable bands. In order to minimize artifactual variation (Black, 1993; Ellsworth et al., 1993), all samples were run at the same time, and repeatability tests were carried out with different DNA template concentrations. The amplification reactions included negative controls, with water instead of template DNA. RAPD-PCR amplified samples were loaded onto 1X TBE 5% glycerol, 5% acrylamide, and 2% N, N-methylenebisacrylamide non-denaturing gel in a 38x50cm Sequi Gen II set apparatus (Bio-Rad, Richmond, California) for 16 h at 25 °C, and 345 volts (13-15 mA), and developed with silver stain (Hiss et al., 1994). Gels were air-dried and bands were photographed, scanned and finally scored directly on the gel. Allele sizes were estimated from the co-migrating molecular size standard, 1 kb molecular ladder (Bethesda Research Laboratory, Gaithersburg, MD).
The binary matrices were processed with the FORTRAN RAPDPLOT program for determining molecular similarity (Kambhampati et al., 1992; Black, 1993), as described by Dergam et al. (1998). An unweighted pair group method with arithmetic averages (UPGMA) was calculated using the PHYLIP 3.5C software package NEIGHBOR option (Felsenstein, 1993), and a phenogram was drawn using the DRAWGRAM option of the same package. For the estimation of Neis (1972) genetic distance among samples, the RAPDDIST program (West & Black, 1998) was used, and statistical support obtained by bootstrapping (Efron, 1985) and CONSENSE (PHYLIP 3.5c), for selection of the best tree. In the bootstrap test, RAPD loci are resampled from the entire data set, and in these new sets some loci are present, while others are not. High bootstrap values therefore reflect the degree of molecular similarity among populations and, thereby, the strength of estimated similarities among them. An Exact Test was carried out to test for allelic heterogeneity (presence/absence) among samples, as implemented in the TFPGA package (http/bioweb.usu.edu/mpmbio/).
For the initial assessment of mtDNA diversity, a 400 bp fragment from 16S ribosomal DNA with a nested primer GGATCTTTTGGTCAGAAK (Dergam, 1996) was amplified together with primer 16 Sar (CGCCTGTTTATCAA AAACAT) (Kocher et al., 1989), corresponding to positions 3172-2958 of the Carassius auratus genome. Amplifications were performed using total genomic DNA, and the polymerase chain reaction (PCR) (Saiki et al., 1988) was done in 50 I of reaction mixture, in PTC-IOO thermal cyclers (MJ Research, Inc., MA), with the following step cycles: 5 min at 95 °C, 2 min at 80 °C, 10 cycles of 1 min at 92 °C, 1 min at 49 °C, 2 min at 72 °C, 32 cycles of 1 min at 92 °C, 35 s at 54 °C, 2 min at 72 °C, and a final extension of 7 min at 72 °C. Haplotype diversity was assessed using Single Strand Conformation Polymorphism (SSCP) (Orita et al., 1989a,b), following the staining protocol described by Hiss et al. (1994). Since the variability of the 16S fragment sequence was known to be too low for populational analyses (Dergam, 1996), 16S SSCP patterns were used as an indirect assessment of haplotype cytb diversity.
Sequencing revealed a 383 bp fragment of cyt b with primers L14725 (CGAAGCTTGATATGAAAAACCAT CGTTG) (Pääbo, 1990) and H15149 (AAACTGCAGCCC TCAGAA TGATATTTGTCCTCA) (Kocher et al., 1989), corresponding to positions 15296-15672 in the Carassius auratus genome. Amplification cycles were: 5 min at 95 °C, 2 min at 80 °C, 10 cycles of 3 min at 92 °C, 1 min at 50 °C, 1:30 min at 72 °C, 32 cycles of 1 min at 92 °C, 35 s at 54 °C, 1:30 min at 72 °C, and a final extension of 7 min at 72 °C. The amplified products were sequenced directly by cycle sequencing (Black and Peisman, 1994), and sequences were read manually from autoradiographs with the SEQAID 113.6 software package (Rhoads and Roufa, 1989); multiple sequences were aligned with CLUSTAL V (Higgins and Sharp, 1989). For maximum parsimony and neighbor joining, Hasegawa, Kishino and Yano (1985) distances were used. Trees were obtained with PAUP 4.0 (Swofford, 2002), with a 2n = 40 chromosome trahira from the Uruguay river basin as outgroup. Uncorrected percent sequence divergence (number of substitutions divided by the total number of bases multiplied by 100) and the number of transitions (TS) and transversions (TV) were estimated for each pair of sequences. Character analyses included different weights applied to third and first positions, and different costs of TS vs. TV.
RESULTS
RAPD variation
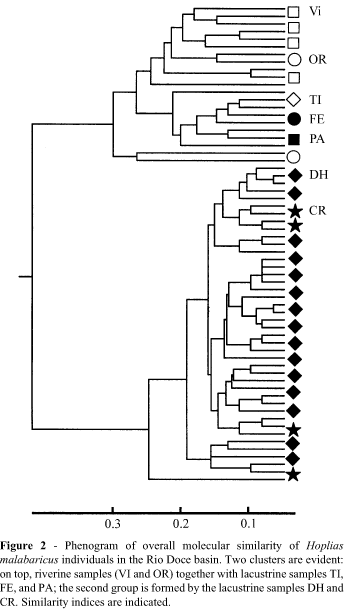
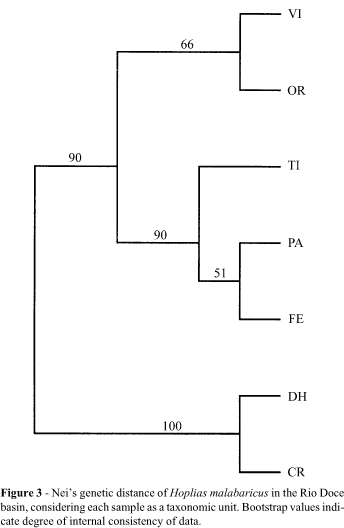
Phenetic (overall similarity) analyses, based on RAPD-PCR fingerprinting, permitted separation of individual fishes into two population sets supported by high bootstrap values (Figure 2). The first group was composed of riverine (Vl and OR) and some lacustrine populations (Tl, FE, and PA), while the second group included the remaining lacustrine populations (DH and CR). The samples displayed from 32 to 38 bands, and a positive correlation was observed between sample size and number of scored bands (r = 0.76). The riverine populations Vl and OR shared a single allele, while fish from Tl, FE and PA lacked diagnostic alleles. The populations DH and CR had two diagnostic alleles, and two other alleles were fixed only in them. When considering each sample as a taxonomic unit, Neis genetic distance yielded clusters which were also supported by high bootstrap values (Figure 3), depicting the same pattern as shown in Figure 2. The allele heterogeneity of populations DH and CR differed significantly from all other populations (Table II).
MtDNA variation
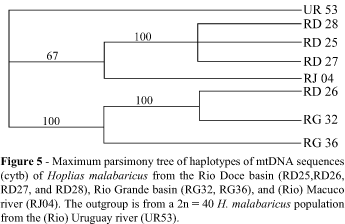
SSCP patterns revealed six putative haplotypes (Figure 4) in the Rio Doce (Genbank accession numbers: RD25 = AF489098; RD26 = AF489099; RD27 = AF489100; RD28 = AF489097) and in the Rio Grande (VG32 = AF489101; VG36 = AF489102). Geographic distribution of mtDNA haplotypes in the Rio Doce basin was independent (p = 0.42) (Table III). All different haplotypes detected by SSCP were sequenced (Figure 4). Table IV shows the percent sequence divergence and number of transitions/ transversions among these sequences and the outgroup (Genbank accession numbers: RJ04 = AF489103; UR53 = AF489104).
The uncorrected sequence divergence was relatively high in the Rio Doce, with a range of 0.3-7.3% and a mean of 3.78%, and the mean divergence between Rio Doce and Rio Macacu sequences was 6.92%. The Rio Grande haplotypes showed a 3.1% divergence from each other. Transitions outnumbered transversions in all pairwise comparisons, with a mean TS/TV of 22 within the Rio Doce. Most of the mutations were in the third position of the codon (93%), while the remaining were restricted to the first position; substitutions were conservative (Irwin et al., 1991). The most generalized sequence in the Rio Doce (RD26) was likewise similar (1%) to one of the sequences of the Rio Grande basin (Figure 5). Maximum parsimony trees with and without weighting of the first position showed the same topology, as was also the case with Neighbor Joining (not shown).
DISCUSSION
RAPD variation
RAPD-PCR markers have been extensively used in populational studies (e.g., Kambhampati et al., 1992), and seem to be efficient for geographic analysis of populations with varying degrees of geographic isolation (e.g., Dergam et al., 1998; Thorpe et al., 1994b). Observed differences between samples may result from selective pressure, historic (vicariant) events, and non-selective processes such as random genetic drift and founder events. However, detection of natural selection in the wild is not an easy task (e.g., Thorpe et al., 1994a) and must be carried out under controlled conditions. Though much still remains to be understood, RAPD-PCR alleles are considered to derive from repetitive regions of the genome (Haymer, 1994), and are thus assumed to be neutral markers.
The authors have interpreted the high molecular similarity of samples CR and DH as the result of a common geologic history of the lakes, probably enhanced by the sedentary nature of H. malabaricus. Multilocus molecular markers are also sensitive to short periods of genetic isolation and, given the low levels of gene flow, populations may accumulate genetic differences in the lapse of relatively few generations (Gravilets and Hastings, 1996). Historic records may shed light on the differential accessibility of the lakes to some exotic and native species. Non-indigenous species always appeared first in lake Dom Helvécio, and only later in lake Carioca. In fact, in 1983, peacock-bass and piranhas were present only in Lake Jacaré (Sunaga and Verani, 1985); in lake Dom Helvécio, both species appeared only in 1985; piranhas did not reach lake Carioca until 1992 (Godinho et aI., 1994), probably via its intermittent connection with the Turvo basin (Figure 1). To date, this lake system has received relatively more attention than other aquatic environments in the basin, and some evidence points toward the existence in this system of two processes: one of differentiation of H. malabaricus populations, and another of migratory movements between lacustrine and riverine populations of other species. Ferreira et al. (1989) reported the occurrence of karyotypic polymorphism in the H. malabaricus population of lake Carioca, where one specimen (out of 17) bore a karyotype with a peculiar configuration of some chromosome pairs, differing from the more widespread karyomorph. In contrast, all H. malabaricus specimens from lake Dos Patos were of the widespread form, which is also present in Viçosa (Dergam, pers. obs.). Menezes (1987) described a new species of characin, Oligosarcus solitarius, in the lake system. The holotype was collected in lake Carioca and is distinguishable from Oligosarcus argenteus by minor morphological characters. Menezes (1988) suggested that O. solitarius might have originated by a vicariant process within the lake system. Vieira (1994) reported that, while most lake Verde fish seemed to complete their life cycle in the lake, one curimatid, Cyphocharax gilberti, apparently completed its reproductive cycle in the river, having usually been caught by fishermen during these migrations.
MtDNA variation
Avise et al. (1987) coined the term phylogeography for the study of geographic patterns of genealogical lineages, including supra- and infraspecific levels of variation, based mainly on the variation of mtDNA patterns. Recently, other biochemical and molecular techniques have also been applied to phylogeographic studies of a large number of organisms (Avise, 1998). Mitochondrial DNA characteristics, such as maternal and clonal patterns of inheritance and high mutation rate, have favored its extensive application to a wide range of biological questions, from studies of high-level systematics to closely related species and populations (Wilson et al., 1985; Avise, 1994). In the last decade, mitochondrial sequencing data have been the preferred approach in phylogeographic studies (Avise, 1994; Avise, 1998). Animal mtDNA is characterized by a much higher number of transitions than transversions between closely related species, with a tendency to level off over increasingly longer times (Brown et al., 1979).
Estimation of time divergence of mtDNA clones based on percentage genetic divergence over time (molecular clock) has been considered as one of the potentially most important contributions of molecular data to the understanding of evolutionary processes (Hillis et al., 1996). It was first proposed by Zuckerkandl and Pauling (1962, 1965), and it is the object of much debate (for a recent review, see Nei and Kumar, 2000). Actual calibration of this clock depends on paleontological and/or geological data (for a review, see Lundberg, 1998). Because of its stochastic nature, molecular clock calibrations are also associated to wide confidence intervals (Hillis et al., 1996). Studies of molecular clock in Neotropical taxa are still scant. Bermingham et al. (1997) calibrated the amount of sequence divergence of 19 geminate species of marine fish, isolated between 2.9 and 3.5 million years ago (mya) by the formation of the Panama Isthmus, and reported an overall rate of 1.2% sequence divergence per million years. In a comprehensive molecular systematics study of serrasalmine characiform genera based on 12S and 16S mitochondrial RNA sequences, Ortí et al. (1996) reported pairwise divergences ranging from 2.8-9.8%. Based on a generalized molecular clock rate of 1-2% sequence divergence per million years, Ortí et al. (1996) estimated the age of divergence of serrasalmine genera as being 1.4-2.8 million. However, the fossil evidence indicates a minimum age of 13.5 million years for genera such as Colossoma and the piranha genera Serrasalmus, Pygocentrus or Prystobrycon (Lundberg, 1998), which, if taken into consideration, would reduce the molecular rate to a low 0.21-0.26% per million years (Lundberg, 1998). In callichthyid fishes, a combination of molecular (again using 12S and 16S ribosomal sequences) and paleontological data results in even lower mutation rates (0.05-0.08% per million years) (Lundberg, 1998). In H. malabaricus, the existence of two haplotypes with a 1% sequence divergence, isolated by vicariant events in the Rio Doce and Rio Grande basins, raises questions about the mutation rate in this species complex. Considering an estimated age of isolation between 12-2 mya (Saadi, 1991), based on the time elapsed since the watershed formation, the mutation rate would be 0.083 - 0.5% per million years, suggesting also maximum lineage splitting ages of 91-5.2 mya within the Rio Doce basin. In this basin, the molecular variation level of H. malabaricus also contrasts with the anostomid Leporinus copelandii, which displays no variation in the same segment of cytb mtDNA (Dergam, unpubl. data). Dergam (1996) reported a much lower sequence divergence level of a 16S ribosomal mt DNA of H. malabaricus, which yielded poorly resolved trees that were also inconsistent with cytogenetic data (Dergam, 1996). The applicability of molecular clock estimates across molecules and Neotropical taxa obviously demands further research.
Although it is accepted that the continent of South America has experienced a process of west-east compression since the Early Cretaceous (118 mya) plate tectonic setting (Lundberg et al., 1998), molecular evidence of more recent vicariant events for Neotropical fish in southeastern Brazil is currently lacking. Today, the Rio Doce and the Rio Grande basins share a common headwater within the Mantiqueira highlands, with a system of mini-grabens and mountain ranges (Saadi, 1991), that may have allowed for the exchange of fish fauna, notably in the Airuoca catchment before the Plio-Pleistocene tectonic uplift of the divide, and hence it is possible that they were connected in the past. On the other hand, it has long been recognized that at least part of the southern basin of the Paraíba do Sul river shares its history with the upper Tietê basin, one of the Paraná river tributaries (for a review, see Malabarba, 1998). Two populations of H. malabaricus from the Paraíba do Sul basin are 2n = 42 (Dergam, unpubl. data), and molecular data of coastal populations from the Guaíba river (Rio Grande do Sul) to the Rio Doce (Minas Gerais) indicate the existence of a well-supported, mainly coastal clade (Dergam, 1996). Thus, cytogenetic and molecular data suggest that the coastal clade of H. malabaricus 2n = 42 entered the Paraná basin, where it is sympatric with H. malabaricus 2n = 39 /40 for males and females respectively (Scavone et al., 1994) and reproductively isolated (Dergam, 1996).
The most parsimonious tree indicates that three of the Rio Doce haplotypes are more related to another coastal basin (RJ) from Rio de Janeiro State. To date, except for some studies of geographic distribution patterns of characins such as Oligosarcus (Menezes, 1987) and some species of Mimagoniates (Weitzman et al., 1987), relatively few studies have addressed the evolutionary implications of coastal distribution of some fish taxa. These authors suggested that at least part of the observed coastal pattern could be explained by geologic events occurred since the last Wisconsin glaciation (60,000 to 16,000 years ago), that lowered the sea level down to 100 m. However, the RJ haplotype shows an average divergence of 7% from the Rio Doce haplotypes, suggesting a much older vicariant event for this species. The close relationship between the most plesiomorphic Rio Doce sequence and one of the Rio Grande sequences suggests a shared history between these basins, corroborating the recent finding of neo-tectonic displacement of the Mantiqueira highlands (Saadi, 1991). It is also possible that the Paraíba do Sul basin may hold haplotypes common to both of the former basins. Langeani (1989) reported a fish fauna in the upper Tietê river similar to the one of the Rio Paraíba do Sul, which was hypothesized as an evidence of an Atlantic drainage of both rivers. Bizerril (1994) concluded that the composition of the coastal fish fauna is best explained by the contribution of the Paraná and São Francisco river basins. Molecular and cytogenetic data of H. malabaricus suggest that coastal basins are, in fact, more related to each other, and that part of the continental biodiversity received a contribution from coastal lineages. While sympatric populations of H. malabaricus have been reported in the Paraná/Plata basins (Dergam, 1996; Dergam et al., 1998; Scavone et al., 1994; Lopes et al., 1998), cytogenetic and molecular data support the view of a single coastal clade of this species complex. The Rio Doce basin also harbors a lower number of species than the Rio Grande and Paraná basins, and may therefore be more prone to the impact of exotic species (Kennedy et al., 2002).
CONCLUSION
The data support the view that, within the Rio Doce basin, RAPD-PCR markers are sensitive to Quaternary events, and molecular similarity between the lacustrine populations DH and CR may indicate that these two lakes share a similar geological history, which is also supported by their highly similar species composition (Sunaga and Verani, 1985). Like other biochemical and molecular markers, techniques based on RAPD alleles do not necessarily involve the genomic regions responsible for adaptive traits which have been traditionally used for granting Evolutionary Significant Units (ESU) status to local populations or species (Allendorf, 1995). However, differences in allele frequencies among populations may also be applied to identify potential management units (Moritz et al., 1995). Based on the data collected, it seems prudent that future policies for non-indigenous species control in the Rio Doce basin should take into account both the management of the water-level of tributaries and the physical isolation of lakes, to protect the native ichthyofauna. If the observed RAPD-PCR genetic pattern for the differentiation of H. malabaricus is also valid for the ichthyofauna of the lake system as a whole, the loss of local populations of some native species constitutes an irreversible process which eroded a portion of the genetic richness of the basin.
Mitochondrial DNA phylogeography shows that the Rio Doce basin and perhaps other coastal populations of H. malabaricus have not evolved in isolation, and that pre-Holocene events are responsible for these complex molecular and cytogenetic findings. Although it has been acknowledged that the distribution patterns of Neotropical fishes do not necessarily follow Quaternary dynamics (Weitzman and Fink, 1983), a combination of RAPD-PCR and mtDNA markers allows to analyze genetic phenomena encompassing different time frames.
ACKNOWLEDGMENTS
The authors wish to thank W.C. Black, IV, for allowing to conduct laboratorial work at Colorado State University, and P. Buckup, R. Secioso, L.G. Cotta and L.W. Assad for collecting samples from the lakes of the Macacu and Doce rivers, as well as Instituto Estadual de Florestas, Companhia Agrícola Florestal-Santa Barbara (CAF) and Centro Mineiro de Conservação da Natureza for providing information, lodging facilities and general assistance. Financial support was provided to J.A.D in part by CAPES (Grant N 2732/92) and Universidade Federal de Viçosa.
Send correspondence to Jorge A. Dergam. Departamento de Biologia Animal, Universidade Federal de Viçosa, 36571-000 Viçosa, MG, Brazil. E-mail: dergam@mail.ufv.br.
- Allendorf FW (1995) Genetics: defining the units of conservation. Am Fish Soc Symp 17:247-248.
- Avise JC, Arnold, J., Ball, R.M., Bermingham E, Lamb T, Neigel JE, Reeb CA and Saunders NC (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Mol Biol Evol 9:475-473.1
- Avise JC (1994) Molecular Markers, Natural History and Evolution. Chapman and Hall, New York.
- Avise JC (1998) The history and purview of phylogeography: a personal reflection. Mol Ecol 7:371-379.
- Black WC IV (1993) PCR with arbitrary primers: approach with care. Insect Mol Biol 2:1-6.
- Black WC IV, and Peisman J (1994) A phylogeny of hard and soft tick taxa based on mitochondrial 16S ribosomal DNA sequences. Proc Natl Acad Sci U.S.A. 91:10034-10038.
- Bizerril CRSF (1994) Análise taxonômica e biogeográfica da ictiofauna de água doce do Leste brasileiro. Acta Biol Leopoldensia 16:51-80.
- Brown WM, Prager EM, Wang A and Wilson AC (1979) Mitochondrial DNA sequences in primates: tempo and mode of evolution. J Mol Evol 18:225-239.
- Dergam JA (1996) Phylogeography and character congruence within the Hoplias malabaricus Bloch, 1794 (Erythrinidae, Characiformes, Ostariophysi) species complex. Ph. D. Dissertation. Colorado State University, Colorado.
- Dergam JA, Suzuki HI, Shibatta OA, Duboc LF, Júlio Jr HF, Caetano-Giuliano L and Black WC IV (1998) Molecular biogeography of the Neotropical fish Hoplias malabaricus (Erythrinidae:Characiformes) in the Iguaçu, Tibagi, and Paraná rivers. Gen Mol Biol 21:493-496.
- Efron B (1985) Bootstrap confidence intervals for a class of parametric problems. Biometrika 72:45-58.
- Ellsworth DL, Rittenhouse KR and Honeycutt RL (1993) Artifactual variation in randomly amplified polymorphic DNA banding patterns. Biotechniques 14:214-217.
- Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) v 3.5c Department of Genetics, University of Seattle, WA.
- Ferreira R, Fonseca CG, Bertollo LAC and Foresti F (1989) Cytogenetics of fishes from Parque Florestal do Rio Doce (MG). I. Preliminary study of Hoplias malabaricus (Pisces, Erythrinidae) from Lagoa Carioca and Lagoa dos Patos. Rev Brasil Genet 12:219-226.
- Godinho AL, da Fonseca MT and de Araújo L (1994) The ecology of predator fish introductions: the case of Rio Doce Valley lakes. In: Pinto-Coelho RM, Giani A and von Sperling E (eds) Ecology and human impact on lakes and reservoirs in Minas Gerais with special reference to future development and management strategies. SEGRAC - Belo Horizonte, MG, Brazil, pp 77-83.
- Godinho AL and Formagio PS (1992) Efeitos da introdução de Cichla ocellaris e Pygocentrus sp. sobre a comunidade de peixes da lagoa Dom Helvécio, MG. In: Associação Mineira de Aquicultura (ed) Anais do X Encontro Anual de Aquicultura, Belo Horizonte, MG, Brazil, pp 93-102.
- Godinho AL and Vieira F (1998) Ictiofauna. In: Costa C et al. (eds) Biodiversidade em Minas Gerais: um atlas para sua conservação. Fundação Biodiversitas, Belo Horizonte, Minas Gerais, Brazil, pp 44-46.
- Gravilets S and Hastings A (1996) Founder effect speciation: a theoretical assesment. Amer Nat 147:466-491.
- Hasegawa M, H Kishino and Yano T (1985) Dating the human-ape split by a molecular clock of mitochondrial DNA. J Mol Evol 22:169-174.
- Haymer DS (1994) Random amplified polymorphic DNAs and microsatellites: what are they, and can they tell us anything we dont really know? Ann Entomol Soc Am 87:717-722.
- Higgins DG and Sharp PM (1989) Fast and sensitive multiple sequence alignments on a microcomputer. Comp Appl Biosc 5:151-153.
- Hillis DM, Mable BK and Moritz C (1996) Application of molecular systematics. In: Hillis DM, Mable BK and Moritz C (eds) Molecular Systematics, Sunderlands, Massachusetts, pp 515-543.
- Hiss RH, Norris DE, Dietrich CR, Whitcomb RF, West DF, Bosio CF, Kambhampati S, Peisman J, Antolin MF and Black WC IV (1994) Molecular taxonomy using single strand conformation polymorphism (SSCP) analysis of mitochondrial DNA genes. Insect Mol Biol 2:171-182.
- Irwin DM, Kocher TD and Wilson AC (1991) Evolution of the cytochrome b in mammals. J Mol Evol 32: 128-144.
- Kambhampati S, Black WC IV and Rai KS (1992) Random amplified polymorphic DNA of mosquito species and populations (Diptera: Culicidae): techniques, statistical analysis, and applications. J Med Entomol 29:939-945.
- Kennedy TA, Naeem S, Howe KM, Knops JMH, Tilman D and Reich P (2002) Biodiversity as a barrier to ecological invasion. Nature 417:636-638.
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX and Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals. Proc Natl Acad Sci USA 86:6196-6200.
- Langeani F (1989) Ictiofauna do alto curso do Tietê (SP): taxonomia. Unpubl. M. Sc. Dissertation. Universidade de São Paulo, São Paulo, 231 p.
- Lopes PA, Alberdi AJ, Dergam JA and Fenocchio AS (1998) Cytotaxonomy of Hoplias malabaricus (Osteicthyes, Erythrinidae) in the Aguapey River (Province of Corrientes, Argentina). Copeia 1998:485-487.
- Lundberg JG (1998) The temporal context for the diversification of Neotropical fishes. In: Malabarba LR et al. (eds) Phylogeny and Classification of Neotropical Fishes. Edipucrs, Porto Alegre, Brasil, pp 49-68.
- Lundberg JG, Marshall L G, Guerrero J, Horton B, Malabarba MCSL and Wesselingh F (1998) The stage for Neotropical fish diversification: a History of tropical South America rivers. In: Malabarba, LR et al. (eds) Phylogeny and Classification of Neotropical Fishes. Edipucrs, Porto Alegre, Brasil, pp 13-48.
- Malabarba CSL (1998) Phylogeny of fossil Characiformes and paleobiography of the Tremembé formation, São Paulo, Brazil. In: Malabarba, LR et al. (eds) Phylogeny and Classification of Neotropical Fishes. Edipucrs, Porto Alegre, Brasil, pp 69-84.
- Menezes NA (1987) Três espécies novas de Oligosarcus Günther, 1864 e redefinição taxonômica das demais espécies do gênero (Osteichthyes, Teleostei, Characidae). Bol. Zool Univ S Paulo 11:1-39.
- Menezes NA (1988) Implications of the distribution patterns of the species of Oligosarcus (Teleostei, Characidae) from central and southern South America. In: Heyer WR and Vanzolini PE (eds) Proceedings of a Workshop on Neotropical Distribution Patterns. Academia Brasileira de Ciências, RJ, Brazil, pp 295-304.
- Moritz C, Lavery S and Slade R (1995) Using allele frequency and phylogeny to define units for conservation and management. Am Fish Soc Symp 17: 249-262.
- Nei M (1972) Genetic distance between populations. Am Nat 106:283-292.
- Nei M and Kumar S (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York, pp 333.
- Orita M, Iwahana H, Kanazawa H, Hayashi K and Sekiya T (1989a) Detection of polymorphisms of human DNA by gel electrophoresis as single strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766-2770.
- Orita M, Suzuki Y, Sekiya T and Hayashi KH (1989b) Rapid and sensitive detection of point mutations and polymorphisms using the polymerase chain reaction. Genomics 5:874-879.
- Pääbo S (1990) Amplifying ancient DNA. In: Innis MA et al., (eds) PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, pp 159-166.
- Petri S and Fúlfaro VJ (1983) Geologia do Brasil. Queiroz TA (ed.). Editora da Universidade de São Paulo, São Paulo, SP, pp 631.
- Rhoads DD and Roufa DJ (1989) SeqAid II Users Manual, version 3.6. Kansas State Univ., Kansas.
- Saadi A (1991) Ensaio sobre a morfotectônica de MG (tensões intra-placa, descontinuidades crustais e morfogênese). Full Professors Dissertation, field of Geomorphology, UFMG, Instituto de Geociências, Belo Horizonte, 1991, 285 pp.
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB and Ehrlich HA (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491.
- Scavone MDP, Bertollo LAC and Cavallini MM (1994) Sympatric occurrence of two karyotypic forms of Hoplias malabaricus (Pisces, Erythrinidae). Cytobios 80:223-227.
- Sunaga T and Verani JR (1985) Preliminary report of comparative study on fish community of the Rio Doce lake. In: Water Research Institute, Limnological Studies in Central Brazil (1st Report). Nagoya University, Chikusa-ku, Nagoya, Japan, pp 167-174.
- Sunaga T and Verani JR (1991)The fish communities of the lakes in the Rio Doce Valley, Northeast Brazil. Verh Internat Verein Limnol 24:2563-2566.
- Swofford D (2002) PAUP* Phylogenetic Analysis Using Parsimony (and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- Thorpe RS, Brown RP, Day M, Malhotra A, McGregor DP and Wüster W (1994a) Testing ecological and phylogenetic hypotheses in microevolutionary studies. In: Eggleton P and Vane-Wright R (eds) Phylogenetics and Ecology. Linnean Society Symposium Series, 17, San Diego, California, USA., pp 189-206.
- Thorpe RS, McGregor DP, Cumming AM and Jordan WC (1994b) Evolution and colonization sequence of island lizards in relation to geological history: mtDNA RFLP, cytochrome b, cytochrome oxidase, 12S rRNA sequence, and nuclear RAPD analysis. Evolution 48:230-240.
- Tundisi JG and De Meis MRM (1985) Geomorphology and limnological processes at the Middle Rio Doce Valley lakes, In: Water Research Institute, Limnological Studies in Central Brazil (1st Report). Nagoya University, Chikusa-ku, Nagoya, Japan, pp 11-17.
- Vari RP and Malabarba LR (1998) Neotropical ichthyology: an overview, In: Malabarba, LR et al. (eds). Phylogeny and Classification of Neotropical Fishes. Edipucrs, Porto Alegre, Brasil, pp 1-11.
- Vieira F (1994) Estrutura de comunidade e aspectos da alimentação e reprodução dos peixes em dois lagos do médio rio Doce, MG. Masters Thesis, Universidade Federal de Minas Gerais, Belo Horizonte.
- Weitzman SH and Fink WL (1983) Relationships of the neon tetras, a group of South American freshwater fishes (Teleostei, Characidae), with comments on the phylogeny of the New World Characiformes. Bull Mus Comp Zool Harv 150:339-365.
- Weitzman S, Menezes NA and Weitzman MJ (1987) Phylogenetic biogeography of the Glandulocaudini (Teleostei: Characiformes, Characidae) with comments on the distributions of other freshwater fishes in eastern and southeastern Brazil, In: Vanzolini PE and Heyer WR (eds) Proceedings of a Workshop on Neotropical distribution patterns. Academia Brasileira de Ciências, Rio de Janeiro, Brazil, pp 379-427.
- West DW and Black WC IV (1998) Breeding structure of three species of Snow Pool Aedes mosquito species in northern Colorado. Heredity 81:371-380.
- Wilson AC, Cann RL, Carr SM, George M Jr, Gyllenstein UB, Helm- Bychowsky KM, Higuchi RG, Palumbi SR, Prager EM, Sage RD and Stoneking M (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biol J Linn Soc 26:375-400.
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA and Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res. 18:6531-6535.
- Zuckerkandl E and Pauling L (1962) Molecular disease, evolution and genetic heterogeneity. In: Kasha M and Pulman B (eds) Horizons in Biochemistry. Academic Press, New York, pp 189-225.
- Zuckerkandl E and Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V and Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97-166.
Publication Dates
-
Publication in this collection
12 Mar 2003 -
Date of issue
2002
History
-
Accepted
27 July 2002 -
Received
Feb 2002