Abstract
A técnica da reação em cadeia pela DNA polimerase (PCR) foi usada para a amplificação rápida de um fragmento de 456bp da região única curta (Us) do genoma do BHV-1. Iniciadores de 18pb do gene da ORF1 foram usados para a amplificação das amostras-padrão e brasileiras. Uma amplificação clássica não foi bem sucedida. A amplificação foi obtida quando se escolheu uma região com baixa concentração de GC no DNA do BHV-1 e através da desnaturação térmica (95° C para 5min) seguida de ciclos térmicos (94° C por 1min e 30seg; 52° C por 1min; 72° C por 1min e 30seg; e ainda por 35 ciclos de 94° C por 1min; 52° C por 1min; 72° C por 1min e 30seg; usando um tempo de extensão final de 72° C por 5min). A clivagem com Pst I confirmou a especificidade do fragmento da ORF 1 do BHV-1. A amplificação do fragmento em todas as amostras testadas sugere que a região é fortemente conservada no genoma do BHV-1. Este PCR poderá detectar rapidamente amostras clínicas, sendo sensível e específico para diagnosticar infecções pelo BHV-1.
Bovino; BHV-1; reação em cadeia pela polimerase; PCR; ORF-1
Bovino; BHV-1; reação em cadeia pela polimerase; PCR; ORF-1
Bovine herpesvirus 1; ORF-1; polymerase chain reaction; Brazil
COMMUNICATION
(Comunicação)
Detection of different Brazilian strains of the bovine herpesvirus-1 (BHV-1) by polymerase chain reaction
(Detecção de amostras brasileiras do herpesvirus bovino 1 (BHV-1) pela reação em cadeia pela polimerase).
A.L.
Cândido
1, A.S.
Martins
2, P.B.
Barros
1, M.
Resende1
* Recebido para publicação, após modificação, em 12 de março de 1999. *Autor para correspondência Autor para correspondência E-mail: mresende@mono.icb.ufmg.br The polymerase chain reaction (PCR) technique first described for primer-directed enzymatic amplification of genomic sequences (Saiki et al., 1985) is now widely used (Innis et al., 1990). Bovine herpesvirus type 1 (BHV-1), a member of the alphaherpesvirinae subfamily (Roizman et al., 1992) is an important pathogen causing serious diseases in cattle. Its more than 70% GC rich genome (Leung-Tack et al., 1994) has not been satisfactory explored by PCR (Vilcek, 1993).
1Depto de Microbiologia, ICB - UFMG
Caixa Postal 486
Av. Antônio Carlos, 6627. Campus Pampulha
31270-901 - Belo Horizonte, MG
2Depto. Fisiologia e Biofisica, ICB UFMG
In a previous report, Leung-Tack et al. (1994) described the complete sequence of the short unique region (Us) of BHV-1 strain ST. Computer analysis of sequences revealed open reading frames (ORFs) whose translation products showed homology in a variety of alphaherpesviruses, except the first open reading frame (ORF1), whose deduced translation product encoded by the complementary strand did not reveal any significant homology with proteins contained in the Swissprot database (Leung-Tack et al., 1994).
In this study, a 456bp ORF1 fragment of the BHV-1 Us genomic region was selected for amplification by PCR. Using the software PCIGENE program for DNA analysis, ORF1 has been shown to be 60% GC enriched with no significant homology, at the nucleic acid level, in other herpesviruses. The synthetic oligonucleotide primers P1-5'-GTACTGGCTC ATGTTTCC-3' (position 430-417) and P2-5-CATGTACCGCCAGGGCAC-3' (position 885-868) were used.
To obtain BHV-1 genomic DNA samples, supernatant of infected MDBK cells was clarified by low speed centrifugation for 20min at 4,000g. Virus was concentrated by ultracentrifugation for 2h at 220,000g and resultant pellet resuspended in TE buffer (10mM Tris-HCl. PH 7.6, 1mM EDTA). Virions were lysed with 1% SDS with 20mg proteinase K/ml and incubated for 2h at 37ºC. DNA was extracted twice with phenol:chlorofom (1:1) and chloroform: isoamylalcohol (24:1) and precipitated with ethanol at 20ºC overnight.
The DNA pellet was collected by centrifugation at 12,000g washed with 70% ethanol, dried and resuspended in TES (Tri-HCl 10mM, pH 8.0, EDTA 1mM, NaCl 10mM).
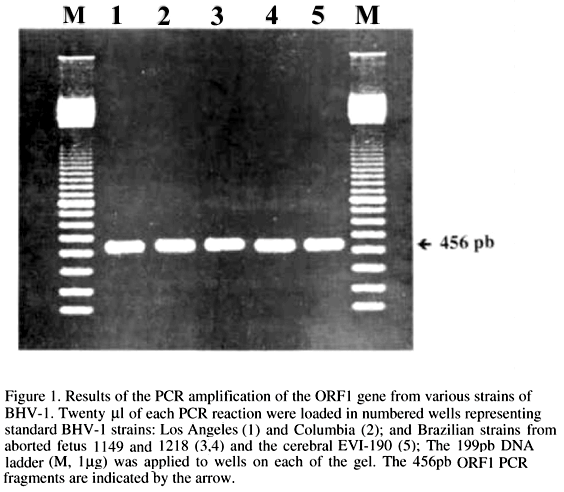
PCR was carried out according to the following protocol: 2ml of each dNTP (dATP 200mM, dGTP 200mM, dCTP 200mM, dTTP 200mM); 10ml of 10´ reaction buffer (Perkin Elmer); 0.5ml (2.5 U) of AmpliTaqRDNA Polymerase; 5ml (50pmol) of P1; 5ml (50pmol) of P2; 10ml (50ng/ml) of 1:10 viral DNA in TES (Tri-HCl 10mM, pH 8.0, EDTA 1mM, NaCl 10mM); the volume completed to 100ml with sterile filtered water and with a 100ml layer of mineral oil. Amplification was carried out in a MJ/Research minicycler by thermal denaturation at 95ºC for 5min prior to 5 cycles of 94ºC for 1,5min; 52ºC for 1min; 72ºC for 1,5min followed by 35 cycles of 94ºC for 1min; 52ºC for 1min; 72ºC for 1,5min; and final extension time of 72ºC for 5min. The amplification products were analysed by electrophoresis in 2% agarose gel (Tris-borate-EDTA buffer, 100 V for 45min) and detected by staining with ethidium bromide (Maniatis et al., 1982). The specificity of the PCR was evaluated with the following viruses: equine herpesvirus 1, equine herpesvirus 4. All viruses were grown in cell culture. Fig. 1 shows the PCR amplification of the 456bp fragments of five selected BHV-1 strains. On the basis of the restriction map analysis of the BHV-1 Us region sequence, by the RESTRI program of the software PC/GENE, the restriction endonuclease Pst I was selected for the digestion and restriction analysis of the amplified fragment. The fragments released by Pst I were estimated by computer analysis as being 300 and 156bp. Fig. 2 shows the confirmation of the BHV-1 ORF1 gene amplification sequence, by cleavage of unpurified PCR products (0.5-1.0mg of DNA) with 10 units of restriction enzyme Pst I (Life Technologis, Inc.)digestion for 2 hours. The Pst I digestion yielded two fragments, as expected.
The results reported here show the possibility of specifically amplify different strains of Brazilian BHV-1.The most likely cause for failure of a PCR amplification is an incomplete denaturation step of the target template that reduces product yield. In contrast, higher or longer denaturation steps lead to loss of enzyme activity (Innis et al., 1990). Major problems with amplification of DNA from certain herpesviruses arise, probably due to their GC rich genomes. The BHV-1 genome is known to be more than 70% GC rich (Leung-Tack et al., 1994), this high GC content also increases the chances of non specific bands (Innis et al., 1990). In order to avoid the amplification of such artifact products of PCR, the selection of a region with lower GC content was strategically important to successfully amplify the BHV-1 DNA fragment. In addition, using the classical amplification protocol for BHV-1 ORF1 gene by PCR (95ºC for 1min 52ºC for 1min 72ºC for 1min) the 456bp fragment could not be detected by means of ethidium bromide stained agarose gels. Thus, for successful amplification and detection, a pre-dwelling temperature of 95ºC for 5min was carried out prior to the first cycle, followed by reduction of denaturation temperature of cycles to 94ºC in order to prolong Taq DNA polymerase life time activity (Yap & McGee, 1991). Under these experimental conditions and after a total of 40 amplification cycles, clear bands of 456bp (Fig. 1) of all BHV-1 strains were detected by agarose-ethidium bromide electrophoresis. Cleavage with Pst I confirmed BHV-1 ORF1 fragment and suggests that Pst I recognition site in the viral genome, flanked by primers P1 and P2, are strongly conserved not only in the published BHV-1 sequence (Leung-Tack et al., 1994) but also in laboratory and field strains of BHV-1 tested in this study. And seems to be a cleavage site pattern characteristic of BHV-1 strains tested.
To our knowledge, the characterization of BHV-1 strains from different clinical sources still remains to be optimised due to considerable homology of the genome sequences among the Herpesviridae family (Leung-Tack et al., 1994). The selected primers and the conditions for PCR reaction of the 456bp fragment BHV-1 (ORF1 gene) indicate that under conditions suggested PCR has a good applicability in the diagnosis of BHV-1 infections of cattle, of aborted fetuses and early identification of viral isolates in tissue culture. While virus isolation in conjunction with a neutralization test require several days, the PCR can be completed within 24 hours.
This present finding suggests this PCR can be applied successfully to clinical samples providing specificity and sensitivity in the detection of BHV-1 infections.
Keywords: Bovine herpesvirus 1, ORF-1, polymerase chain reaction, Brazil
ACKNOWLEDGMENTS
This work was supported by Programa de Apoio ao Desenvolvimento Científico e Tecnológico (PADCT II), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Pró-Reitoria de Pesquisa (PRPq-UFMG) and Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG). We wish to thank Prof. Rômulo C. Leite (Escola de Veterinária-UFMG), for his kind gift of the 1218 and 1149 strains. The BHV-1 EVI-190 strain was gently given by Dr. Paulo M. Roehe, Instituto de Pesquisa Veterinária Desidério Finamor (IPVDF-RS). This research has been submitted by A.L. Cândido in partial fulfillment for the MS in Microbiology at the Instituto de Ciências Biológicas - UFMG, Belo Horizonte, Brazil.
RESUMO
A técnica da reação em cadeia pela DNA polimerase (PCR) foi usada para a amplificação rápida de um fragmento de 456bp da região única curta (Us) do genoma do BHV-1. Iniciadores de 18pb do gene da ORF1 foram usados para a amplificação das amostras-padrão e brasileiras. Uma amplificação clássica não foi bem sucedida. A amplificação foi obtida quando se escolheu uma região com baixa concentração de GC no DNA do BHV-1 e através da desnaturação térmica (95° C para 5min) seguida de ciclos térmicos (94° C por 1min e 30seg; 52° C por 1min; 72° C por 1min e 30seg; e ainda por 35 ciclos de 94° C por 1min; 52° C por 1min; 72° C por 1min e 30seg; usando um tempo de extensão final de 72° C por 5min). A clivagem com Pst I confirmou a especificidade do fragmento da ORF 1 do BHV-1. A amplificação do fragmento em todas as amostras testadas sugere que a região é fortemente conservada no genoma do BHV-1. Este PCR poderá detectar rapidamente amostras clínicas, sendo sensível e específico para diagnosticar infecções pelo BHV-1.
Palavras-chaves: Bovino, BHV-1, reação em cadeia pela polimerase, PCR, ORF-1
- INNIS, M.A., GELFAND, D.H., SNINSKV, J.J. et al. PCR protocols: a guide to methods and applications San Diego: Academic, 1990. 354p.
- LEUNG-TACK, P., AUDONNET, J., RIVIERE, M. The complete DNA sequence and the genetic organization of the short unique region (Us) of the herpesvirus type 1 (ST Strain). J. Microbiol., v.2, p.409-421, 1994.
- MANIATIS T, FRITSCH E.F., SAMBROOK J. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor, 1982. 545p.
- ROIZMAN, B., DESROSIERS, R.C. FLECKENSTEIN, B. et al. The family Herpesviridae: An update. Arch. Virol, v.123, p.425-449, 1992.
- SAIKI, R.K., SHARF, S., FALOONA, F. et al.. Enzymatic amplification of b-globin genomic sequences and restriction site analysis for the diagnosis of sickle-cell anemia. Science, v.230, p.1350-1354, 1985.
- VILCEK, S. Detection of the bovine herpesvirus-1 (BHV-1) genome by PCR. J. Virol. Meth., v.41, p.245-248, 1993.
- YAP, E.P.H., McGEE, O'D.J. Short PCR product yields improved by lower denaturation temperatures. Nucl. Acids Res., v.19, p.1713-1717, 1991.
Publication Dates
-
Publication in this collection
19 Apr 2001 -
Date of issue
June 1999
History
-
Received
12 Mar 1999