Abstract
BACKGROUND
Leishmaniasis is a neglected tropical disease caused by the parasite Leishmania braziliensis, commonly found in Brazil and associated with cutaneous and visceral forms of this disease. Like other organisms, L. braziliensis has an enzyme called glutamine synthetase (LbGS) that acts on the synthesis of glutamine from glutamate. This enzyme plays an essential role in the metabolism of these parasites and can be a potential therapeutic target for treating this disease.
OBJECTIVES
Investigate LbGS structure and generate structural models of the protein.
METHODS
We use the method of crosslinking mass spectrometry (XLMS) and generate structural models in silico using I-TASSER.
FINDINGS
42 XLs peptides were identified, of which 37 are explained in a monomeric model with the other five indicating LbGS dimerization and pentamers interaction region. The comparison of 3D models generated in the presence and absence of XLMS restrictions probed the benefits of modeling with XLMS highlighting the inappropriate folding due to the absence of spatial restrictions.
MAIN CONCLUSIONS
In conclusion, we disclose the conservation of the active site and interface regions, but also unique features of LbGS showing the potential of XLMS to probe structural information and explore new drugs.
Key words:
structural proteomics; protein; glutamine synthetase; Leishmania braziliensis
According to the Centers for Disease Control and Prevention (CDC), leishmaniasis comprises a group of neglected tropical diseases caused by parasites of the genus Leishmania. This disease presents itself in three forms: cutaneous (CL), mucous (ML), and visceral (VL). Cutaneous leishmaniasis can cause substantial morbidity, while visceral leishmaniasis can be fatal.11. CDC - Centers for Disease Control and Prevention. Parasites - Leishmaniasis [Internet]. Centers for Disease Control and Prevention. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
https://www.who.int/news-room/fact-sheet...
,22. WHO - World Health Organization. Leishmaniasis [Internet]. World Health Organization. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
https://www.who.int/news-room/fact-sheet...
In Brazil, data from 2019 show 2,529 new cases of VL, with 1.2 cases per 100,000 inhabitants and a mortality rate of 9%. For CL, 15,484 new cases were confirmed (7.37 cases per 100,000 inhabitants), of which 67.1% had clinical cure 1.9% abandoned treatment and 19 deaths.33. MS/SVS - Ministério da Saúde/Secretaria de Vigilância em Saúde. Doenças tropicais negligenciadas: 30 de janeiro - Dia mundial de combate às doenças tropicais negligenciadas. Boletim Epidemiológico. Número especial. 2021. Available from: https://www.gov.br/saude/pt-br/media/pdf/2021/marco/3/boletim_especial_doencas_negligenciadas.pdf.
https://www.gov.br/saude/pt-br/media/pdf...
There are at least eight species of Leishmania that can infect humans and lead to the development of parasitosis in Brazil: L. (V.) braziliensis, L. guyanensis, L. (L.) amazonensis, L. (L.) infantum (syn. chagasi) L. (V.) lainsoni, L (V.) naiffi, L. (V.) shawi and L. (V.) lindenbergi.44. Anversa L, Tiburcio MGS, Richini-Pereira VB, Ramirez LE. Human leishmaniasis in Brazil: a general review. Rev Assoc Med Bras. 2018; 64: 281-9. Considering that Leishmaniasis is found on every continent except Australia, Pacific Islands and Antarctica, and there are about 90 countries classified as endemic, WHO estimates approximately 1.2 million new cases of cutaneous leishmaniasis per year.22. WHO - World Health Organization. Leishmaniasis [Internet]. World Health Organization. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
https://www.who.int/news-room/fact-sheet...
For visceral leishmaniasis, it is estimated that the new incidences are currently below 100,000 per year.22. WHO - World Health Organization. Leishmaniasis [Internet]. World Health Organization. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
https://www.who.int/news-room/fact-sheet...
The enzyme glutamine synthetase (GS) is essential in nitrogen metabolism, being responsible for the catalysis of glutamine from ATP, glutamate, and ammonia; this process occurs in two stages, starting with the activation of an intermediate gamma-glutamyl phosphate (γ-G-P), followed by a nucleophilic attack of ammonia in this intermediate releasing phosphate and forming glutamine.55. Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. Structure-function relationships of glutamine synthetases. Biochim Biophys Acta. 2000; 1477: 122-45.,66. Wedler FC, Boyer PD. Substrate binding and reaction intermediates of glutamine synthetase (Escherichia coli W) as studied by isotope exchanges. J Biol Chem. 1972; 247: 984-92.,77. Wedler FC, Horn BR. Catalytic mechanisms of glutamine synthetase enzymes. Studies with analogs of possible intermediates and transition states. J Biol Chem. 1976; 251: 7530-8.,88. Woolfolk CA, Stadtman ER. Regulation of glutamine synthetase. IV. Reversible dissociation and inactivation of glutamine synthetase from Escherichia coli by the concerted action of EDTA and urea. Arch Biochem Biophys. 1967; 122: 174-89. It is found in all organisms, including Leishmania sp, presenting three types: GS-I, found in most prokaryotes, GS-II, found in eukaryotes, and GS-III, found in some prokaryotes.99. Llorca O, Betti M, González JM, Valencia A, Márquez AJ, Valpuesta JM. The three-dimensional structure of an eukaryotic glutamine synthetase: functional implications of its oligomeric structure. J Struct Biol. 2006; 156: 469-79. The GS types I and II are dodecamers formed by two hexameric rings maintained mainly by hydrophobic interactions. The GS type III is formed by two hexameric rings associate across opposite interfaces, each ring has flipped 180º with respect to its position in the other two types.1010. Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28.,1111. van Rooyen JM, Abratt VR, Belrhali H, Sewell T. Crystal structure of type III glutamine synthetase: surprising reversal of the inter-ring interface. Structure. 2011; 19: 471-83. Glutamine synthesis sequence of L. braziliensis (LbGS) is formed by two pentameric rings interacting, probably, by hydrophobic interactions due to the conservation (in relation to HsGS) of the sequence rich in prolines and lysines. Hydrogen bonds and salt bridges sustain the interaction of monomers, being interface weaker in LbGS than HsGS.1212. Moreira C, Ramos M, Fernandes P. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21: 1028.
MATERIALS AND METHODS
In this work, the nucleotide sequence encoding (GenBank CAM36993.1) the putative LbGS was cloned into a pET28a plasmid vector. A 42.35 kDa protein was obtained by overexpressing LbGS in the Escherichia coli (DE3) NiCo strain with 1 mM IPTG at 30ºC for four hours. The recombinant protein was purified from the soluble fraction of cellular lysate using a HisTrap column in the Akta Purifier system (GE Healthcare) using buffer A (Sodium phosphate buffer pH 7.4 10 mM, 500 mM NaCl, 40 mM imidazole) to equilibrate the column and a linear gradient of buffer B (pH 7.4 10 mM sodium phosphate buffer, 500 mM NaCl, 1 M imidazole) for elution.
We performed crosslinking experiments (XL) using the purified protein as previously described.1313. Pereira MBM, Santos AM, Gonçalves DC, Cardoso AC, Consonni SR, Gozzo FC, et al. aB-crystallin interacts with and prevents stress-activated proteolysis of focal adhesion kinase by calpain in cardiomyocytes. Nat Commun. 2014; 5: 5159. The protein was digested with trypsin in the proportion of 1/50 (E/S) for 20 hours and the enzymatic reaction and was stopped by adding trifluoroacetic (0.4% v/v final). Subsequently, the peptides were quantified using the fluorometric test - Qubit 4.0® (Invitrogen) according to the manufacturer’s recommendations. Each sample was desalted and concentrated using Stage-Tips (STop and Go-Extraction TIPs) according to literature.1414. Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003; 75: 663-70. The peptide mixture was suspended in 0.1% formic acid and analysed as follows. An Ultimate 3000 (Thermo Fisher®) coupled online with a Fusion Lumos Orbitrap mass spectrometer (Thermo Fisher®) was used for generating the mass spectra data. The peptide mixture was chromatographically separated on a column (15 cm in length with a 75 μm I.D.) packed in-house with ReproSil-Pur C18-AQ 3 μm resin (Dr Maisch GmbH HPLC) with a flow of 250 nL/min from 5% to 50% ACN in 0.1% formic acid in a 140 min gradient. The Fusion Lumos Orbitrap was set to the data-dependent acquisition (DDA) mode to automatically switch between full-scan MS and MS/MS acquisition with 60s dynamic exclusion. Survey scans (200-1500 m/z) were acquired in the Orbitrap system with a resolution of 120,000 at m/z 200. The most intense ions captured in a 2s cycle time were selected, excluding those unassigned and in a 1+ charge state, sequentially isolated and HCD (Higher-energy collisional dissociation) fragmented using a normalised collision energy of 30. The fragment ions were analysed with a resolution of 30,000 at 200 m/z. The general mass spectrometric conditions were as follows: 2.5 kV spray voltage, no sheath or auxiliary gas flow, heated capillary temperature of 250ºC, predictive automatic gain control (AGC) enabled, and an S-lens RF level of 40%. Mass spectrometer scan functions and nLC solvent gradients were controlled by the Xcalibur 4.1 data system (Thermo Fisher®). Protein identification was performed using Pattern Lab for proteomics V available at http://www.patternlabforproteomics.org and a database containing 8,084 sequences of L. braziliensis downloaded from Uniprot. Results were filtered as described in the software’s bioinformatics protocol1515. Carvalho PC, Lima DB, Leprevost FV, Santos MDM, Fischer JSG, Aquino PF, et al. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat Protoc. 2016; 11: 102-17. and only the protein of interest was identified, thus achieving 0% FDR. XL identification was performed with the Spectrum Identification Machine for Cross-Linked Peptides (SIM-XL) software that is freely available at http://www.patternlabforproteomics.org/sim-xl.1616. Lima DB, de Lima TB, Balbuena TS, Neves-Ferreira AGC, Barbosa VC, Gozzo FC, et al. SIM-XL: a powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015; 129: 51-5. The LbGS sequence from L. braziliensis was downloaded on March 29th, 2021, from the NCBI. The search parameters considered: fully tryptic peptide candidates with masses between 600 and 4800 Da, 20 ppm for precursor and fragment mass. The modifications were carbamidomethylation of cysteine and oxidation of methionine as fixed and variable, respectively. The files are available in proteomics.fiocruz.br/LbGS (Supplementary data). The distance of 11.4 Å between cross-linked lysines identified using SIM-XL (scores limit of 1.5 for intralinks and 2.0 for interlink)1616. Lima DB, de Lima TB, Balbuena TS, Neves-Ferreira AGC, Barbosa VC, Gozzo FC, et al. SIM-XL: a powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015; 129: 51-5. was used as an input for I-TASSER.1717. Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015; 43: W174-181. Structural analysis and visual inspection were conducted with EBI-PISA,1818. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007; 372: 774-97. Pymol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger), Wincoot,1919. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004; 60: 2126-32. and ChimeraX.2020. Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30: 70-82.
RESULTS AND DISCUSSION
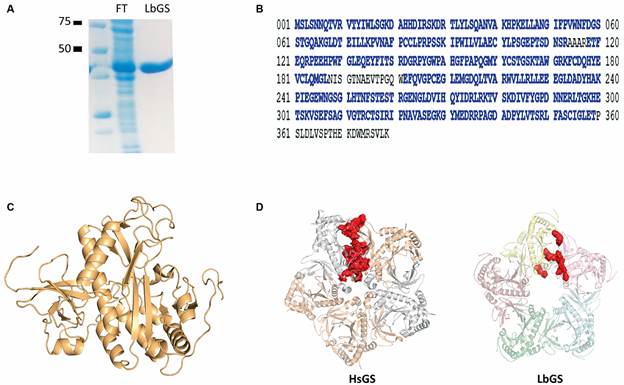
The identity of purified protein (Fig. 1A) was confirmed by mass spectrometry (Fig. 1B). The experimental constraints obtained by XLMS are listed in Table I. The tertiary model (Fig. 1C) displayed a C-score value of 0.89 and a TM-score of 0.83 ± 0.08, which indicates good confidence and correct fold (TM-score > 0.5 suggests a correct fold).1717. Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015; 43: W174-181.,2121. Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004; 57: 702-10. 37 out of 42 XL distances could be placed in the monomeric model, with acceptable distances between 11.4 Å and 35 Å (Table I).2222. Merkley ED, Cort JR, Adkins JN. Cross-linking and mass spectrometry methodologies to facilitate structural biology: finding a path through the maze. J Struct Funct Genomics. 2013; 14: 77-90.
(A) purified recombinant glutamine synthetase (LbGS) (0.4 µg/µL). FT indicates the flow through of chromatography. (B) LbGS sequence with peptides identified by mass spectrometry in blue. (C) the tertiary model obtained with XLMS. (D) pentameric rings from HsGS (PDB ID 2OJW) and LbGS. In red the interacting interface residues.
All GS are oligomers and eucaryotes GS type II are decamers composed of pentameric rings superimposed with monomers comprised of ~ 350 to 420 residues. GSs have ten active sites per oligomer placed in the interface of two interacting monomers.55. Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. Structure-function relationships of glutamine synthetases. Biochim Biophys Acta. 2000; 1477: 122-45.,2323. Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28. The 5 XL restraints that cannot be justified by a monomeric model should indicate the region of dimerization; these results corroborate that, like other GS, the dimerization occurs from the C-terminal region of one monomer and N-terminal of the other (Table I).
The GS decamers present two main interaction interfaces: intra-ring forming the pentamers rings and tail-to-tail between superimposed pentameric rings. The human GS intra-ring interface is formed by the interaction of the N- and C-terminal of two subunits.2424. Moreira C, Ramos MJ, Fernandes PA. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21(8): 1028. Comparing LbGS and HsGS interfaces (Fig. 1D, Tables II-III), we observe a highly conserved C-terminal region but a divergent N-terminal (Fig. 2A). LbGS lacks an N-terminal a-helix which diminishes the size and strength of that interface (1537.4 Å2 and ΔG of -4.3 for LbGS and 2249.1 Å2 and ΔG of -15.4 for HsGS). The number of hydrogen bonds and salt bridges in HsGS are 37 and 16, respectively (Table II). In LbGS, the numbers are quite lower: seven hydrogen bonds and nine salt bridges (Table III). Although the interface of LbGS is less stable than HsGS, the following XLs might indicate that LbGS dimerization occurs in solution: K167-K166, S02-K240, D29-K28, C162-S163, K167-S166, D314-S317, D234-K240. Regarding the tail-to-tail interface, XL residues S163-K167 and S166-K167 can indicate the presence of this interface in our sample.
Studies have shown that the interaction of the pentameric HsGS model also depends on residues L139 to P160, which form a loop rich in proline and glycine, favoring hydrophobic interactions within pentamers.2424. Moreira C, Ramos MJ, Fernandes PA. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21(8): 1028. This loop (I138 to M159, LbGS numbering) is conserved in the LbGS structure and is also rich in prolines and glycines (Fig. 2B). However, the differences in protein sequence result in an β-sheet (R143, R144 and P145, LbGS numbering) not conserved in the homolog HsGS (Fig. 2B).
(A) superposing of glutamine synthetase (LbGS) and HsGS with differences highlighted. LbGS in red, HsGS in blue. (B) LbGS monomeric model with the region from L139 to P160 highlighted (green). Dark green indicates proline and glycine residues. Arrows indicate an β-sheet not conserved in the homolog HsGS.
The active site of GSs comprises three regions: one for glutamate, one for ATP and one for ammonia, with very conserved residues of two subunits of the pentameric ring.2323. Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28. Each monomer is divided into two domains, each contributing with the active site of the adjacent monomer: N-terminal (smaller) is composed of a sheet formed by six antiparallel β-strands which two take parts of the active site; C-terminal (the larger), formed mainly by α-helix and six β-strands formed by most of the residues that make up the active site99. Llorca O, Betti M, González JM, Valencia A, Márquez AJ, Valpuesta JM. The three-dimensional structure of an eukaryotic glutamine synthetase: functional implications of its oligomeric structure. J Struct Biol. 2006; 156: 469-79. (Fig. 3A). In the model obtained by the crosslinking method, the active site residues E133, E135, N247, G248, H252, R294, R314, E333 and R335 (Glutamate site, LbGS numbering) (Fig. 3B), G186, S256 and R319 (ATP site, LbGS numbering) (Fig. 3C) are fully conserved together with the ammonia site, which involves three residues from the C-terminal region (E195, E202 and E300, LbGS numbering) and two residues from the N-terminal region (D58 and S60, LbGS numbering) from the adjacent subunit (Fig. 3D). The structural active sites differences found by superposing the GsHS and LbGS structures reside in: (i) glutamate site from 287-303 (LbGS numbering, Fig. 2A), while R299 (PDB_ID 2OJW), which is the terminal part of a loop and R294 (LbGS) which is part of an α -helix; (ii) the ammonia site from 55-63, being D63 (PDB_ID 2OJW) parts of a β-sheet, and D58 (LbGS), part of a loop (Fig. 2A).
the active site of glutamine synthetase (LbGS). (A) in pentamer, each monomer is divided into two domains N-terminal (blue) and C-terminal (blue). (B) the glutamate site. (C) the ammonia site. (D) ATP site. Numbers references subunits of the pentameric ring.
superposing of glutamine synthetase (LbGS) models generated with and without XL restrictions. The divergent regions S17-D24, N38-P53, G288-E306 from LbGS modeled with (blue) and without (red) restrictions are highlighted.
Finally, we also predicted a model using only the sequence of LbGS without XL restrictions. The model constructed by I-TASSER displayed good confidence scores (C-score value of -0.12, TM-score of 0.70 ± 0.12). When both LbGS models (modeled with and without XL restrictions) are superimposed, they present an RMSD of 0.671Å and the following regions would be modeled inappropriately in the absence of the restraints obtained from experimental XL: S17-D24, N38-P53, G288-E306 (Fig. 4). Thus, it is relevant to say that spatial restrictions give modeling a sense of the real conformation of the enzyme in vitro, differently from the model generated only based on homology allowing us to evaluate their in-solution conformation, in addition to the comparison with already known 3D counterparts structures available in the PDB. The technique of drug design aims both small therapeutic molecules targeting protein as itself as drug (biotherapeutics). Knowing proteins conformation in vitro and the ligand binding sites are the heart of structure-based drug development. Currently, this strategy depends not only on structural information, but also on dynamics, kinetics, and enzyme-substrate interaction data, that together provide the dynamic information on protein’s in vitro conformation and flexibility and are possible due to the computational advances that emerged over years.2525. Renaud J. Structural biology in drug discovery: methods, techniques, and practices. 1st ed. Wiley. 2020. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9781118681121.
https://onlinelibrary.wiley.com/doi/book...
,2626. Renaud J. The evolving role of structural biology in drug discovery. In: Renaud J, org. Structural biology in drug discovery. 1st ed. Wiley. 2020. p. 1-22. Available from: https://onlinelibrary.wiley.com/doi/10.1002/9781118681121.ch1.
https://onlinelibrary.wiley.com/doi/10.1...
Some studies have used GS enzymes as a therapeutic target to treat diseases such as cancer, malaria, and leishmaniases.2323. Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28.,2424. Moreira C, Ramos MJ, Fernandes PA. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21(8): 1028.,2727. Berlicki L. Inhibitors of glutamine synthetase and their potential application in medicine. Mini Rev Med Chem. 2008; 8: 869-78.,2828. Singh N, Siddiqi MI. Computational evaluation of glutamine synthetase as drug target against infectious diseases: molecular modeling, substrate-binding analysis, and molecular dynamics simulation studies. Med Chem Res. 2017; 26: 450-60.
The LbGS lacks structural and functional studies being the studies with GS from L. donovani (LdGS) the closest to LbGS. Kumar et al performed biochemical studies that demonstrate the enzyme’s dependence on divalent metals for its optimal activity and optimum pHs from 7 to 9, similar to HsGS.2929. Kumar V, Yadav S, Soumya N, Kumar R, Babu NK, Singh S. Biochemical and inhibition studies of glutamine synthetase from Leishmania donovani. Microb Pathog. 2017; 107: 164-74. Also, the provide a structural comparison of LdGS and HsGS describing relevant non-conserved residues for substrate recognition (E7, L132, S190, S249 and V205, LdGS numbering) and the importance of the electropositive potential in the active sites.3030. Kumar V, Sushma Sri N, Tripathi N, Sharma VK, Bharatam PV, Garg P, et al. Structural exploration of glutamine synthetase from Leishmania donovani: insights from in silico and in vitro analysis. Int J Biol Macromol. 2020; 146: 860-74. These differences allowed them to find specific LdGS inhibitors, that might act in LbGS as we observed that the residues are conserved in LbGS. The potential of GS from Leishmania sp as therapeutic target was also evidenced by knock-out experiments indicating the dependence of parasite proliferation and infectivity on external supply of glutamine.3131. Kumar V, Ghosh S, Roy K, Pal C, Singh S. Deletion of glutamine synthetase gene disrupts the survivability and infectivity of Leishmania donovani. Front Cell Infect Microbiol. 2021; 11: 622266.
Herein, we provide LbGS structural investigation identifying the active site, important interfaces, and unique structural features from LbGS. All these information allow investigation for new drugs.
REFERENCES
-
1CDC - Centers for Disease Control and Prevention. Parasites - Leishmaniasis [Internet]. Centers for Disease Control and Prevention. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
» https://www.who.int/news-room/fact-sheets/detail/leishmaniasis -
2WHO - World Health Organization. Leishmaniasis [Internet]. World Health Organization. 2020. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
» https://www.who.int/news-room/fact-sheets/detail/leishmaniasis -
3MS/SVS - Ministério da Saúde/Secretaria de Vigilância em Saúde. Doenças tropicais negligenciadas: 30 de janeiro - Dia mundial de combate às doenças tropicais negligenciadas. Boletim Epidemiológico. Número especial. 2021. Available from: https://www.gov.br/saude/pt-br/media/pdf/2021/marco/3/boletim_especial_doencas_negligenciadas.pdf
» https://www.gov.br/saude/pt-br/media/pdf/2021/marco/3/boletim_especial_doencas_negligenciadas.pdf -
4Anversa L, Tiburcio MGS, Richini-Pereira VB, Ramirez LE. Human leishmaniasis in Brazil: a general review. Rev Assoc Med Bras. 2018; 64: 281-9.
-
5Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. Structure-function relationships of glutamine synthetases. Biochim Biophys Acta. 2000; 1477: 122-45.
-
6Wedler FC, Boyer PD. Substrate binding and reaction intermediates of glutamine synthetase (Escherichia coli W) as studied by isotope exchanges. J Biol Chem. 1972; 247: 984-92.
-
7Wedler FC, Horn BR. Catalytic mechanisms of glutamine synthetase enzymes. Studies with analogs of possible intermediates and transition states. J Biol Chem. 1976; 251: 7530-8.
-
8Woolfolk CA, Stadtman ER. Regulation of glutamine synthetase. IV. Reversible dissociation and inactivation of glutamine synthetase from Escherichia coli by the concerted action of EDTA and urea. Arch Biochem Biophys. 1967; 122: 174-89.
-
9Llorca O, Betti M, González JM, Valencia A, Márquez AJ, Valpuesta JM. The three-dimensional structure of an eukaryotic glutamine synthetase: functional implications of its oligomeric structure. J Struct Biol. 2006; 156: 469-79.
-
10Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28.
-
11van Rooyen JM, Abratt VR, Belrhali H, Sewell T. Crystal structure of type III glutamine synthetase: surprising reversal of the inter-ring interface. Structure. 2011; 19: 471-83.
-
12Moreira C, Ramos M, Fernandes P. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21: 1028.
-
13Pereira MBM, Santos AM, Gonçalves DC, Cardoso AC, Consonni SR, Gozzo FC, et al. aB-crystallin interacts with and prevents stress-activated proteolysis of focal adhesion kinase by calpain in cardiomyocytes. Nat Commun. 2014; 5: 5159.
-
14Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003; 75: 663-70.
-
15Carvalho PC, Lima DB, Leprevost FV, Santos MDM, Fischer JSG, Aquino PF, et al. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat Protoc. 2016; 11: 102-17.
-
16Lima DB, de Lima TB, Balbuena TS, Neves-Ferreira AGC, Barbosa VC, Gozzo FC, et al. SIM-XL: a powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015; 129: 51-5.
-
17Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015; 43: W174-181.
-
18Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007; 372: 774-97.
-
19Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004; 60: 2126-32.
-
20Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30: 70-82.
-
21Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004; 57: 702-10.
-
22Merkley ED, Cort JR, Adkins JN. Cross-linking and mass spectrometry methodologies to facilitate structural biology: finding a path through the maze. J Struct Funct Genomics. 2013; 14: 77-90.
-
23Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008; 375: 217-28.
-
24Moreira C, Ramos MJ, Fernandes PA. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets. Molecules. 2016; 21(8): 1028.
-
25Renaud J. Structural biology in drug discovery: methods, techniques, and practices. 1st ed. Wiley. 2020. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9781118681121
» https://onlinelibrary.wiley.com/doi/book/10.1002/9781118681121 -
26Renaud J. The evolving role of structural biology in drug discovery. In: Renaud J, org. Structural biology in drug discovery. 1st ed. Wiley. 2020. p. 1-22. Available from: https://onlinelibrary.wiley.com/doi/10.1002/9781118681121.ch1
» https://onlinelibrary.wiley.com/doi/10.1002/9781118681121.ch1 -
27Berlicki L. Inhibitors of glutamine synthetase and their potential application in medicine. Mini Rev Med Chem. 2008; 8: 869-78.
-
28Singh N, Siddiqi MI. Computational evaluation of glutamine synthetase as drug target against infectious diseases: molecular modeling, substrate-binding analysis, and molecular dynamics simulation studies. Med Chem Res. 2017; 26: 450-60.
-
29Kumar V, Yadav S, Soumya N, Kumar R, Babu NK, Singh S. Biochemical and inhibition studies of glutamine synthetase from Leishmania donovani. Microb Pathog. 2017; 107: 164-74.
-
30Kumar V, Sushma Sri N, Tripathi N, Sharma VK, Bharatam PV, Garg P, et al. Structural exploration of glutamine synthetase from Leishmania donovani: insights from in silico and in vitro analysis. Int J Biol Macromol. 2020; 146: 860-74.
-
31Kumar V, Ghosh S, Roy K, Pal C, Singh S. Deletion of glutamine synthetase gene disrupts the survivability and infectivity of Leishmania donovani. Front Cell Infect Microbiol. 2021; 11: 622266.
Publication Dates
-
Publication in this collection
10 Jan 2022 -
Date of issue
2021
History
-
Received
14 June 2021 -
Accepted
26 Oct 2021