Abstracts
This study discusses from the theoretical standpoint the analyses of heterosis and combining ability from the partial diallel. Aiming at to investigate the information provided by the genetic parameters, simulation studies were realized. The genetic model adjusted considered one gene, groups of six populations, three degrees of dominance (|d/a| equal to 0.5, 1 and 2, representing partial, complete and overdominance) and random allelic frequencies. For each degree of dominance 50 replications were performed. The model for heterosis analysis does not need to be restricted. The unrestricted model gives the same information as the restricted one. The model for combining ability is restricted. The restrictions of the partial-G2 model of GERALDI and MIRANDA-FILHO do not satisfy the parametric values of the specific combining ability effects. As in the complete diallel analysis, from the theoretical point of view there is no differences between the analysis of heterosis and combining ability. The analysis of differences in gene frequencies among the populations of the two groups is more efficient when it considers the heterosis and not the specific heterosis - or the specific heterosis effects or the effects of specific combining ability - mainly for genes whose frequencies are contrasting in the groups.
average heterosis; variety heterosis; specific heterosis; general combining ability; specific combining ability
Este estudo discute do ponto de vista teórico as análises de heterose e capacidade de combinação a partir de dialelo parcial. Foram realizados estudos de simulação com o objetivo de investigar as informações proporcionadas pelos parâmetros genéticos. O modelo genético ajustado considerou um gene, grupos de seis populações, três graus de dominância (|d/a| igual a 0,5, 1 e 2, representando dominâncias parcial e completa, e sobredominância) e freqüências alélicas aleatórias. Para cada grau de dominância foram realizadas 50 simulações. O modelo para análise de heterose não precisa ser restrito. O modelo irrestrito proporciona as mesmas informações do restrito. O modelo para análise de capacidade de combinação é restrito. As restrições do modelo partial-G2 de GERALDI e MIRANDA-FILHO não satisfazem aos valores paramétricos dos efeitos de capacidade específica de combinação. Como na análise de dialelo completo, do ponto de vista teórico não há diferenças entre as análises de heterose e de capacidade de combinação. A análise das diferenças de freqüências gênicas entre as populações dos dois grupos é mais eficiente quando são consideradas as heteroses e não as heteroses específicas - ou os efeitos de heterose específica ou os efeitos de capacidade específica de combinação - principalmente quanto a genes cujas freqüências são contrastantes nos grupos.
heterose média; heterose varietal; heterose específica; capacidade geral de combinação; capacidade específica de combinação
PLANT BREEDING
Heterosis and combining ability analyses from the partial diallel
Análises de heterose e capacidade de combinação com dialelo parcial
José Marcelo Soriano Viana* * autor correspondente.
Universidade Federal de Viçosa, Departamento de Biologia Geral, 36570-000 Viçosa (MG). E-mail: jmsviana@ufv.br
ABSTRACT
This study discusses from the theoretical standpoint the analyses of heterosis and combining ability from the partial diallel. Aiming at to investigate the information provided by the genetic parameters, simulation studies were realized. The genetic model adjusted considered one gene, groups of six populations, three degrees of dominance (|d/a| equal to 0.5, 1 and 2, representing partial, complete and overdominance) and random allelic frequencies. For each degree of dominance 50 replications were performed. The model for heterosis analysis does not need to be restricted. The unrestricted model gives the same information as the restricted one. The model for combining ability is restricted. The restrictions of the partial-G2 model of GERALDI and MIRANDA-FILHO do not satisfy the parametric values of the specific combining ability effects. As in the complete diallel analysis, from the theoretical point of view there is no differences between the analysis of heterosis and combining ability. The analysis of differences in gene frequencies among the populations of the two groups is more efficient when it considers the heterosis and not the specific heterosis - or the specific heterosis effects or the effects of specific combining ability - mainly for genes whose frequencies are contrasting in the groups.
Key words: average heterosis, variety heterosis, specific heterosis, general combining ability, specific combining ability.
RESUMO
Este estudo discute do ponto de vista teórico as análises de heterose e capacidade de combinação a partir de dialelo parcial. Foram realizados estudos de simulação com o objetivo de investigar as informações proporcionadas pelos parâmetros genéticos. O modelo genético ajustado considerou um gene, grupos de seis populações, três graus de dominância (|d/a| igual a 0,5, 1 e 2, representando dominâncias parcial e completa, e sobredominância) e freqüências alélicas aleatórias. Para cada grau de dominância foram realizadas 50 simulações. O modelo para análise de heterose não precisa ser restrito. O modelo irrestrito proporciona as mesmas informações do restrito. O modelo para análise de capacidade de combinação é restrito. As restrições do modelo partial-G2 de GERALDI e MIRANDA-FILHO não satisfazem aos valores paramétricos dos efeitos de capacidade específica de combinação. Como na análise de dialelo completo, do ponto de vista teórico não há diferenças entre as análises de heterose e de capacidade de combinação. A análise das diferenças de freqüências gênicas entre as populações dos dois grupos é mais eficiente quando são consideradas as heteroses e não as heteroses específicas - ou os efeitos de heterose específica ou os efeitos de capacidade específica de combinação - principalmente quanto a genes cujas freqüências são contrastantes nos grupos.
Palavras-chave: heterose média, heterose varietal, heterose específica, capacidade geral de combinação, capacidade específica de combinação.
1. INTRODUCTION
The partial diallel genetic design was suggested by COMSTOCK and ROBINSON (1948) to estimate components of genotypic variance in cross-pollinating populations. Although no relevant theoretical contribution was added, KEMPTORNE and CURNOW (1961) demonstrated that it is a suitable design for this purpose and could be more efficient than others. This objective, however, has almost always been attained by using design I and its variations, because of better fit to the alogamous plants reproductive system.
The partial diallel is almost exclusively used for analysis of heterosis and/or combining ability of cross-pollinating populations, inbred families (topcross with more than one tester), inbred lines or pure lines, but not plants. Although it is common to consider the partial diallel as an alternative to the diallel when the number of parents is high (GERALDI and MIRANDA-FILHO, 1988; MIRANDA-FILHO and GERALDI, 1984), it should be the choice of the breeder when there are two different population groups, such as corn populations adapted to tropical climate and exotic populations, adapted to temperate climate; S3 families, and broad and/or narrow genetic base testers; dent and flint inbred lines; and common bean varieties tolerant and not tolerant to cold, and when there is no interest in hybrids of populations of the same group.
The methodologies of MIRANDA-FILHO and GERALDI (1984), for heterosis analysis, and of GERALDIi and MIRANDA-FILHO (1988), for combining ability analysis, are modifications of the models of GARDNER and EBERHART (1966) and GRIFFING (1954), but there is no discussion of the theoretical inferences that may be established. The objective of this study was to discuss heterosis and combining ability analysis from the partial diallel.
2. METHODS
2.1 Heterosis analysis
The genotypic mean of a population in group 1 is
where for each locus, m is the mean of the genotypic values of the homozygotes, pj is the frequency of the gene that increases the trait expression in the population, a is the difference between the genotypic value of the homozygote pj of greatest expression and m, and d is the deviation due to dominance (HALLAUER and MIRANDA-FILHO, 1988). The constant µ is the sum of the mean m for each locus and the parameter vj is the population effect.
The genotypic mean of a population in group 2 is
where for each locus, rj' is the frequency of the gene that increases the trait expression in the population and vj' is the population effect.
The genotypic mean of the hybrid of populations j and j' is
The means of groups 1 and 2 are
where
1 and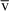 2 are the means of the population effects of groups 1 and 2.
2 are the means of the population effects of groups 1 and 2.
The heterosis manifested in the hybrid of populations j and j' is
The mean of the heterosis manifested in the hybrids of population j of group 1 is
The mean of the heterosis manifested in the hybrids of population j' of group 2 is
The average heterosis is
where
is the mean of the hybrids and
is the arithmetic mean of the group means.
The mean of the hybrid of populations j and j' can be expressed as follows
But
where
is the specific heterosis.
Thus
The mean phenotypic values of populations j and j' and of the hybrid of these populations are
where
j,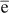 j´ and
j´ and 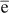 jj are the residues.
jj are the residues.
These equations are not those of the model defined by MIRANDA-FILHO and GERALDI (1984), because they characterize the unrestricted model.
The means of populations j in group 1 and j' in group 2 can be expressed as
where v* represents the population effect expressed as deviation of the mean effect of the group.
Thus, as  ,
,
where  and
and  ´ are the effects of varietal heterosis and
´ are the effects of varietal heterosis and  is the effect of specific heterosis.
is the effect of specific heterosis.
Expressing the population means as function of the arithmetic mean of the group means results
where
 .
.
Thus, the equations of the restricted model presented by MIRANDA FILHO and GERALDI (1984) are
The n1 + n2 + 3 linearly independent parametric restrictions of this model are
 for all j, and
for all j, and  for all j'
for all j'
2.1.1. Hypotheses tests
The most relevant hypotheses that can be tested in the analysis of variance are:
Testing H0(1) and H0(2) is equivalent to testing the equality of the gene frequencies in the populations in the group, that is, that there are no differences of gene frequencies among the populations of the group (H0(1):pjj = pi for all j and, H0(2):Rjj´= rifor all j'). The sum of squares attributable to these hypotheses are associated to (n1 - 1) and (n2 - 1) degrees of freedom. Testing H0(3) is the equivalent to testing that the mean gene frequencies are equal in the two groups (H0(3): =
=  ). The sum of squares due to this hypothesis is associated to one degree of freedom. In the case of rejection of H0(1) and/or H0(2) and/or H0(3) , testing H0(4) is equivalent to testing that there is no dominance (H0(4):di = 0 for all i). One degree of freedom is associated with the sum of squares due to this hypothesis. Absence of dominance can be tested by testing the hypothesis H0:Hjj´= H for all j and j' (unrestricted model) or H0:
). The sum of squares due to this hypothesis is associated to one degree of freedom. In the case of rejection of H0(1) and/or H0(2) and/or H0(3) , testing H0(4) is equivalent to testing that there is no dominance (H0(4):di = 0 for all i). One degree of freedom is associated with the sum of squares due to this hypothesis. Absence of dominance can be tested by testing the hypothesis H0:Hjj´= H for all j and j' (unrestricted model) or H0: = Hjj - H = 0 for all j and j' (restricted model), with the advantage that the sum of squares attributable to this hypothesis (Heterosis sum of squares) is associated to more degrees of freedom (n1n2).
= Hjj - H = 0 for all j and j' (restricted model), with the advantage that the sum of squares attributable to this hypothesis (Heterosis sum of squares) is associated to more degrees of freedom (n1n2).
If there is dominance, testing H0(5) and H0(6) is equivalent to testing H0(1) and H0(2). Although there are differences regarding the corresponding sums of squares, the degrees of freedom are the same. Testing H0(7)is equivalent to testing that there are no differences of gene frequencies among the populations in each group. The sum of squares attributable to this hypothesis is associated to (n1 - 1)(n2 - 1) degrees of freedom.
2.2 Combining ability analysis
The effect of general combining ability of a population in group 1 is
For a population in group 2
Thus
where sjj' is the effect of the specific combining ability of populations j and j'.
The genotypic means of populations j of group 1 and j' of group 2 are
where sjj and sj'j'are the effects of specific combining ability of the population with itself.
Expressing the means of the populations and the hybrid as function of M. results
Therefore, the phenotypic means are
which define a restricted model different from that presented by GERALDI and MIRANDA-FILHO (1988). The n1 + n2 + 3 linearly independent parametric restrictions are
 for all j, and
for all j, and
 for all j'
for all j'
The equivalent model to that defined as partial-G2 by GERALDI and MIRANDA-FILHO (1988) is obtained expressing the genotypic means of the parents and the hybrids as function of the general mean
Thus
It should be pointed out that the restrictions of the partial-G2 model,
 for all j and
for all j and  for all j'
for all j'
do not satisfy the parametric values of the specific combining ability effects.
2.2.1 Hypotheses tests
The most relevant tests in the analysis of variance are:
The hypotheses H0(8) and H0(9) are equivalent to the hypothesis H0(1) and H0(2)in the heterosis analysis, including the degrees of freedom. In the case of rejection of H0(8) and/or H0(9) and/or H0(3), testing hypothesis H0(10) is equivalent to testing absence of dominance. The sum of squares due to this hypothesis (specific combining ability sum of squares) is associated to (n1n2 - 1) degrees of freedom.
The orthogonal decomposition of the sum of squares attributable to the hypothesis H0(10) allows testing the hypotheses
The tests of these hypotheses are equivalent to the tests of hypotheses H0(5), H0(6) and H0(7) in the heterosis analysis. The sum of squares due to hypothesis H0(7) is the sum of squares attributable to hypothesis H0(13).
Therefore, as pointed out by VIANA (2000a, b), relative to the complete diallel, the heterosis and combining ability analyses from the partial diallel are, from the theoretical point of view, redundant and not complementary.
Aiming to investigate the information provided by the genetic parameters of the heterosis and combining ability analyses from the partial diallel, simulation studies were realized. The genetic model adjusted considered one gene, groups of six populations, three degrees of dominance (|d/a| equal to 0.5, 1 and 2, representing partial, complete and overdominance) and random allelic frequencies. For each degree of dominance 50 replications were performed.
3. RESULTS AND DISCUSSION
3.1 Simulations
The population effect is an indicator of the superiority of the population in terms of favorable gene frequencies, regardless of the group. If the populations are pure lines or inbred lines, the superiority is for the number of favorable genes. Based on the simulation, the correlation between the frequency of the gene that increases the trait expression and the population effect varied from 0.7806 to 0.9981, and the mean value was 0.9537 (Table 1).
The value is 1.0 when there is no dominance. Because the population effect cannot be estimated in the unrestricted model, the populations can be classified based on the differences between the population effects, since the contrasts are estimable functions, or more simply, by analysis of the estimates of the population genotypic means, that are also estimable, because the difference between the population mean and the population effect is a constant.
Even fitting the unrestricted model, the heterosis, the average heterosis, the varietal heterosis and the specific heterosis are estimable functions because they are linear combinations of estimable functions, the population and hybrid means. The average heterosis is an indicator of the direction of dominance if there are differences of gene frequencies among the populations of at least one group or if there are differences in the mean gene frequencies of the groups. The dominance direction can also be assessed by sign of the heterosis or the variety and specific heterosis. If the variety heterosis are predominantly positive and the specific heterosis are predominantly negative the dominance is unidirectional positive. The opposite indicates negative dominance.
The varietal heterosis, in absolute value, indicates divergence of the population, in terms of gene frequencies, compared to the mean frequencies in the other group of parents. The simulation study indicated that the correlation between varietal heterosis and the absolute value of the difference between the gene frequency in the population and the mean frequency in the other group of parents varied from 0.6574 to 0.9867, and the mean was 0.9338 (Table 1).
Although the specific heterosis is an indicator of differences of gene frequencies among the populations, it is less efficient than heterosis. The results from simulation showed that, regardless of the degree of dominance, it is better to assess the divergence among the populations by the absolute value of the heterosis. The correlation between the absolute value of the difference of gene frequencies between two populations and the heterosis varied from 0.9263 to 0.9808, and the mean was 0.9604. The minimum, maximum and mean values for specific heterosis were 0.0118, 0.9604 and 0.7077. Regardless of the degree of dominance, the correlation values for specific heterosis were always lower (Table 1). Therefore, the greater the magnitude of the heterosis, the greater the differences of gene frequencies among the populations, and the greater the specific heterosis (respecting the sign), the greater the differences of gene frequencies.
It can be considered that the efficiency of the specific heterosis to indicate differences of gene frequencies among the populations is greater in the case of complete diallel. From the simulation study, it was ascertained that the correlation between specific heterosis and heterosis varied from 0.6743 to 0.9282, with mean value of 0.8198, in the case of the complete diallel. With partial diallel the correlation varied from 0.2085 to 0.9389, with mean value of 0.6894. Regarding the genes whose frequencies are contrasting in the two groups, the correlation between specific heterosis and the absolute value of the difference of gene frequencies among the populations is nil, regardless of the degree of dominance (Table 1).
When the specific heterosis is equal to the mean value (
..= - 2H), the gene frequencies in each population are equal to the mean gene frequencies of the group to which it belongs.From the theoretical point of view, the inferences that can be established from the fit of the restricted model are almost the same as those from the unrestricted model. One difference is that the population effect expressed as deviation of the mean effect of the group is an indicator of the superiority of the population only for the others in the group. In the simulation study the correlation between the frequency of the gene that increases the trait expression and the population effect expressed as deviation of the mean effect of the group varied from 0.6248 to 0.9999, being 1.0 in the absence of dominance (Table 1). Another difference is that the deviations  ,
,  ´and
´and  should be analyzed respecting magnitude and sign and cannot be used to assess the dominance direction.
should be analyzed respecting magnitude and sign and cannot be used to assess the dominance direction.
The general combining ability effect is an indicator of the superiority of the population, in terms of frequency of the favorable genes, and of the differences between the gene frequencies of the population and the mean frequencies in the group. The correlation between the frequency of the gene that increases the trait expression and the effect of general combining ability is 1.0 or -1.0, in the last case only with overdominance and depending on the gene frequencies. A negative value occurred once in the 50 simulations carried out for overdominance (Table 1). When g = 0, the gene frequencies in the population are equal to the mean frequencies in the group. The analysis of the general combining ability effects of the populations in one group permits the same inferences from the analysis of the population effects expressed as deviations from the mean value (v*) and from the varietal heterosis effects (H*) because g = (1/2)v*+H*.
The specific combining ability effect of a population with itself is an indicator, similar to the varietal heterosis, of the differences between gene frequencies in the population and the mean frequencies in the other group of parents. In the case of unidirectional positive dominance, the lower the value (negative), the greater the differences. With negative unidirectional dominance, the greater the value (positive), the greater the differences. In the simulation study, the absolute minimum, maximum and mean values of the correlation between the effect of specific combining ability of the population with itself and the difference between the gene frequency in the population and the mean frequency in the other group of parents were 0.6574, 0.9867 and 0.9338 (Table 1). As Sjj´=Sjj´-(-2H) =  there is no difference between analyzing specific combining ability effect, specific heterosis effect or specific heterosis.
there is no difference between analyzing specific combining ability effect, specific heterosis effect or specific heterosis.
For a more efficient assessment of the differences of gene frequencies among the populations the heterosis should be analyzed, given by
To highlight the similarities and differences with the models of MIRANDA-FILHO and GERALDI (1984) and GERALDI and MIRANDA-FILHO (1988) the data analyzed are the same of the articles.
3.2 Heterosis analysis
Regarding the analysis of variance, the difference between the unrestricted and restricted models is in the sum of squares of group 1 versus group 2 (Table 2). The decomposition of the sum of squares of populations is orthogonal in the unrestricted model. The sums of squares of group 1, of group 2 and of the contrast between the means of the groups are those from the partial-ST model of GERALDI and MIRANDA-FILHO (1988). The others are from the model of MIRANDA-FILHO and GERALDI (1984). The main evidences are that there are differences of gene frequencies only among the populations in group 2, that there are differences between the mean gene frequencies of the groups and that there is dominance.
It should be emphasized the consistency of the tests regarding population effects and those for varietal heterosis or variety heterosis effects.
In relation to the estimates of estimable functions and of their standard deviations, differences were ascertained in the unrestricted and restricted models only in the estimated standard deviation values (Tables 3 and 4). Interestingly, the values estimated by the model of MIRANDA-FILHO and GERALDI (1984) are those obtained by fitting the unrestricted model. The main inferences are: (i) the population with the greatest frequencies of the genes that increase yield is 4'; (ii) there is positive dominance; (iii) the population of group 2 more divergent compared to group 1 is 1'; and (iv) the most divergent populations are 1 and 1'.
3.3 Combining ability analysis
Regarding the analysis of variance, the decomposition of the sum of squares of treatments is non-orthogonal (Table 2). In spite of the differences in the sums of squares corresponding to the models presented by GERALDI and MIRANDA-FILHO (1988), the inferences are exactly the same: there are differences of gene frequencies among the populations in each group, the mean frequencies of the groups are identical and there is positive dominance. The decomposition of the sum of squares of the specific combining ability also showed differences of gene frequencies among the populations in the same group. It is interesting to note that the hypothesis that the specific combining ability effects of the populations of the two groups are equal to zero is not rejected, showing little divergence between them, a result corroborated by the test on the differences of the mean gene frequencies of the groups.
The analysis of the effects of general and specific combining ability and of the heterosis indicated that populations 1 and 1' are superior and the most divergent between each other and compared to the other group of parents (Table 3). Regarding the partial-ST model of GERALDI and MIRANDA-FILHO (1988), there are differences only in the standard deviations of the general combining ability and the specific combining ability of the populations in the two groups (Table 4). Compared to the partial-G2 model, only regarding the effects of specific combining ability of different populations the linear associations of the estimates are not very high. The correlations between estimates of the general combining ability effects of populations of group 1, group 2 , specific combining ability effects of the populations in group 1, group 2, and of the specific combining ability effects of the populations of the two groups, are 0.98, 0.88, 1.0, 1.0 and 0.68, respectively.
Recebido para publicação em 5 de setembro de 2005 e aceito em 29 de agosto de 2006.
- COMSTOCK, R.E.; ROBINSON, H.F. The components of genetic variance in populations. Biometrics, Washington, v.4, p.254-266, 1948.
- GERALDI, I.O.; MIRANDA-FILHO, J.B. Adapted models for the analysis of combining ability of varieties in partial diallel crosses. Brazilian Journal of Genetics, Ribeirão Preto, v.11, p.419-430, 1988.
- HALLAUER, A.R.; MIRANDA FILHO, J.B. Quantitative genetics in maize breeding. 2.ed. Ames: Iowa State University Press, 1988. 468p.
- KEMPTHORNE, O.; CURNOW, R.N. The partial diallel cross. Biometrics, Washington, v.17, p.229-250, 1961.
- MIRANDA-FILHO, J.B.; GERALDI, I.O. An adapted model for the analysis of partial diallel cross. Brazilian Journal of Genetics, Ribeirão Preto, v.7, p.677-688, 1984.
- VIANA, J.M.S. The parametric restrictions of the Gardner and Eberhart diallel analysis model: heterosis analysis. Genetics and Molecular Biology, Ribeirão Preto, v.23, p.869-875, 2000a.
- VIANA, J.M.S. The parametric restrictions of the Griffing diallel analysis model: combining ability analysis. Genetics and Molecular Biology, Ribeirão Preto, v.23, p.877-881, 2000b.
Publication Dates
-
Publication in this collection
12 Apr 2007 -
Date of issue
2007
History
-
Accepted
29 Aug 2006 -
Received
05 Sept 2005






































