Abstracts
The aim of this study was to investigate the presence of the Hepatitis G Virus on a population of blood donors from São Paulo, Brazil and to evaluate its association to sociodemographic variables. Two RT-PCR systems targeting the putative 5'NCR and NS3 regions were employed and the former has shown a higher sensitivity. The observed prevalence of HGV-RNA on 545 blood donors was 9.7% (CI 95% 7.4;12.5). Statistical analysis depicted an association with race/ethnicity, black and mulatto donors being more frequently infected; and also with years of education, less educated donors presenting higher prevalences. No association was observed with other sociodemographic parameters as age, gender, place of birth and of residence. DNA sequencing of nine randomly chosen isolates demonstrated the presence of genotypes 1, 2 and 3 among our population but clustering of these Brazilian isolates was not detected upon phylogenetic analysis.
Hepatitis G Virus; HGV; Blood donors; PCR
O objetivo deste estudo foi investigar a presença do vírus da hepatite G e sua associação com variáveis sóciodemográficas em uma população de doadores de sangue da cidade de São Paulo, Brasil. Dois sistemas de PCR foram empregados, um focando a região 5' não-codificadora e outro a região não-estrutural 3, apresentando o primeiro maior sensibilidade. A prevalência detectada do HGV-RNA em 545 doadores foi de 9,7% (IC 95% 7,4;12,5). A prevalência do HGV-RNA foi significativamente maior em doadores da raça negra/mulata (26,5%) quando comparados aos doadores caucasianos (7,5%), (OR = 4,5). Também foi observada uma tendência de maiores prevalências em doadores de menor escolaridade. Não se observou associação significativa com outros parâmetros estudados como idade, sexo, origem geográfica e local de residência. O sequenciamento do DNA de nove isolados selecionados ao acaso demonstrou a presença dos genótipos 1, 2 e 3, mas não houve um agrupamento das amostras brasileiras quando submetidas à análise filogenética
High prevalence of GB Virus C/Hepatitis G Virus RNA among Brazilian blood donors
Alta prevalência do RNA do vírus da hepatite G (HGV) em doadores de sangue brasileiros
José Eduardo LeviI, II, IV; Daniela Gazoto ContriII; Liliam Pereira LimaI; Deise Tihe TakaokaII; Regina Helena GarriniII; Wellingtom SantosII; Roberta FachiniI, II; Silvano WendelI, II, III
IHospital Sírio-Libanês Blood Bank, São Paulo, SP, Brazil
IICentro de Imunologia e Imunogenética, São Paulo, SP, Brazil
IIIPost-Graduate Student, Infectious Diseases Department, Faculdade de Medicina da Universidade de São Paulo (FMUSP), São Paulo, SP, Brazil
IVVirology Lab., Instituto de Medicina Tropical da Universidade de São Paulo, São Paulo, SP, Brazil
Correspondence Correspondence Dr. Silvano Wendel Rua Adma Jafet 91 01308-050 São Paulo, SP, Brazil Phone: + 55-11-3255-7746. Fax: + 55-11-3257-1290 E-mail : snwendel@uninet.com.br
SUMMARY
The aim of this study was to investigate the presence of the Hepatitis G Virus on a population of blood donors from São Paulo, Brazil and to evaluate its association to sociodemographic variables. Two RT-PCR systems targeting the putative 5'NCR and NS3 regions were employed and the former has shown a higher sensitivity.
The observed prevalence of HGV-RNA on 545 blood donors was 9.7% (CI 95% 7.4;12.5). Statistical analysis depicted an association with race/ethnicity, black and mulatto donors being more frequently infected; and also with years of education, less educated donors presenting higher prevalences. No association was observed with other sociodemographic parameters as age, gender, place of birth and of residence.
DNA sequencing of nine randomly chosen isolates demonstrated the presence of genotypes 1, 2 and 3 among our population but clustering of these Brazilian isolates was not detected upon phylogenetic analysis.
Keywords: Hepatitis G Virus; HGV; Blood donors; PCR.
RESUMO
O objetivo deste estudo foi investigar a presença do vírus da hepatite G e sua associação com variáveis sóciodemográficas em uma população de doadores de sangue da cidade de São Paulo, Brasil. Dois sistemas de PCR foram empregados, um focando a região 5' não-codificadora e outro a região não-estrutural 3, apresentando o primeiro maior sensibilidade.
A prevalência detectada do HGV-RNA em 545 doadores foi de 9,7% (IC 95% 7,4;12,5). A prevalência do HGV-RNA foi significativamente maior em doadores da raça negra/mulata (26,5%) quando comparados aos doadores caucasianos (7,5%), (OR = 4,5). Também foi observada uma tendência de maiores prevalências em doadores de menor escolaridade. Não se observou associação significativa com outros parâmetros estudados como idade, sexo, origem geográfica e local de residência.
O sequenciamento do DNA de nove isolados selecionados ao acaso demonstrou a presença dos genótipos 1, 2 e 3, mas não houve um agrupamento das amostras brasileiras quando submetidas à análise filogenética.
INTRODUCTION
Hepatitis G Virus is a recently identified single-stranded RNA virus that has been detected on blood transfusion recipients and further on other groups presenting parenteral risk factors. It has been classified into the Flaviviridae family sharing a similar genomic arrangement to other members including the Hepatitis C Virus (HCV). The worldwide prevalence of HGV on healthy populations is notably high, ranging from 2% on German blood donors15 to 14.2% on West African residents5. Sequence analysis has allowed the classification of HGV isolates on types and this approach revealed the existence of at least 4 genotypes. Genotype 1 is detected mainly on African residents, genotype 2 is the most prevalent on Europe and North America and genotypes 3 and 4 in Asia10,11. A fifth South African genotype has also been proposed18. Though HGV transmission by blood transfusion is firmly established1, several lines of evidence (reviewed in KLEINMAN7) led to the conclusion that HGV does not cause any form of liver disease, precluding the need of screening in the blood transfusion setting. The aim of this study was to investigate the presence of the Hepatitis G Virus on a population of blood donors from São Paulo, Brazil and to evaluate its association to sociodemographic variables.
METHODS
Study population: Serum samples were obtained from 545 consecutive voluntary blood donors from the Hospital Sírio-Libanês Blood Bank (from August to November of 1997), including 380 males and 165 females, ranging from 18 to 59 years of age. All samples were routinely assessed for serological markers, namely; anti-HIV1/2, HBsAg, anti-HBc, anti-HCV, anti-HTLV-I/II, ALT levels, Syphilis and Chagas disease. Demographic data, including age, gender, marital status, race, local of birth and residence and educational background were obtained from donors by self-assessment.
HGV RT-PCR:RNA was extracted from 250 µL of serum by the use of TRI Reagent LS (GIBCO BRL, São Paulo, Brazil) in accordance with the manufacturer instructions. Total isolated RNA was submitted to reverse transcription with Random Primers 2.5 µM (GIBCO BRL, São Paulo, Brazil) and 50 units of Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT, GIBCO BRL, São Paulo, Brazil) in a volume of 20 µL, for 30 minutes at 37 0C followed by 5 minutes at 95 °C. The putative HGV 5' non-coding region was amplified from 5.0 µL of cDNA, corresponding to 62.5 µL of serum, in a single-round PCR, using antisense primer ( 5' CAC TGG GTG CAA GCC CCA GAA 3'), sense primer ( 5' CAC TGG TCC TTG TCA ACT CGC 3')9 and one unit of Taq DNA Polymerase (GIBCO BRL, São Paulo, Brazil) in a final volume of 25 µL. Denaturation at 95 °C for 5 minutes was followed by 40 cycles of amplification at 94 °C for 30s, 55 °C for 30s and 72 °C for 1 minute. On positive samples, a PCR product of 367 bp was detectable after electrophoresis on an ethidium bromide stained, 1.8% agarose gel.
Samples were also submitted to amplification of a distinct HGV genomic region, the NS3 gene, presumably coding for a viral helicase/protease, using a nested PCR protocol12. For the first round, 2.5 µL of cDNA was submitted to PCR using antisense primer 1 (5' CGA AGT TTC CTG TGT ACC C 3'), sense primer 1 (5' GAC GTT GGT GAG ATC CCC TT 3') in a final volume of 12.5 µL. For the second round 5.0 µL of first round PCR product was amplified using antisense primer 2 (5' TAG AGA GCG CGT CTG TCG CAC 3') and sense primer 2 (5' TGG GCA TGG TAT CCC CCT CGA GC 3') in a final volume of 25 µL. For both rounds, denaturation at 95 °C for 5 minutes was followed by 40 cycles of 94 °C for 45s, 53 °C for 45s and 72 °C for 75s. 238 bp and 196 bp were respectively first and second round PCR products detectable after eletrophoresis on an ethidium bromide stained, 1.8% agarose gel. Positive controls consisted of a HGV positive sample derived from a chronic hepatitis C patient and a plasmid containing HGV cloned sequences gently provided by Dr Jason Pickering (Imperial College, Dept. of Medicine, London, UK). Negative controls consisted of plasma supernatant derived from a donation negative for all serological markers plus HCV-RNA and HGV-RNA by the methods described above.
Sequencing:PCR products from 9 randomly chosen isolates (HGV 5' non-coding region PCR products) were purified on an ultrafiltration system MICROCON - 100 (AMICON, INC., Beverly, USA) and subjected to direct sequencing by using ABI PRISM Dye terminator Cycle Sequencing Ready reaction kit (Perkin-Elmer, Foster City, USA). Sequences were determined by using automated sequencer ABI 377 (ABI, Foster City, USA) equipment at the DNA Sequencing Core - University of Michigan.
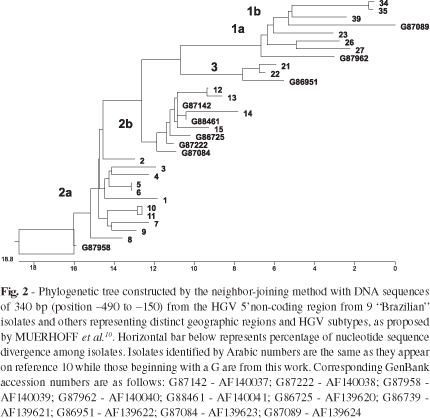
Phylogenetic Analysis: Similarity of the isolates here described to other HGV sequences was verified by a GenBank searching algorithm (BLAST version 2.0 program), available through the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/). DNA sequences representative of distinct HGV groups were obtained from the GenBank and aligned with the sequences here reported by using CLUSTAL-W17 software (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html). An unrooted phylogenetic tree was constructed based on the neighbor-joining analysis of the sequence distance data by using the NEIGHBOR program of the PHYLIP package available through the Web site (ftp://evolution.genetics.washington.edu/pub/phylip).
Statistical Analysis: The c2 or c2 for trend tests were used for investigating the association between two categorical variables. The odds ratio (OR) for the presence of HGV-RNA considering as exposure several sociodemographic variables was estimated by the method of Mantel-Haenzel6 with confidence intervals of 95%.
RESULTS
HGV-RNA was detected on 53 out of 545 serum samples (9.7%, 95% CI 7.4;12.5), by at least one of the two primers sets. Among the HGV infected individuals, there was no reactivity to the following serological markers: HBsAg, anti-HCV, Syphilis, Chagas disease, HIVp24Ag and anti-HIV 1/2 (Table 1). Of the 53 reactive samples, 47 and 30 were respectively reactive to 5'non-coding region and to NS-3 primer pairs while 24 were reactive for both (Table 2). There were no significant associations between presence of HGV-RNA with gender (p = 0.20) nor age (p = 0.15). The relative frequency of HGV-RNA was higher on donors of Negro/Mulatto race (26.5%) than on Caucasians (7.5%) and Orientals (0%) (Fig. 1) (OR = 4.5, 95% CI 2.3;8.5, Table 3). DNA sequences comparison revealed, on all cases, more than 90% of sequence identity to other HGV isolates previously described and phylogenetic analysis doesn't show a cluster of Brazilian samples (Fig. 2).
DISCUSSION
We have found a very high prevalence of HGV-RNA in our blood donor population, corroborating the data from BASSIT et al.3on a blood donor population from another hospital in São Paulo. A prevalence of this magnitude has only been reported, to our knowledge, on West African residents and South African blood donors4,5, except for parenterally exposed groups where even higher prevalences are observed4,15. The finding of this virus in that proportion of blood donors with no correlation to other markers points to the little pathogenicity of this agent, if any. However, the determinants of these high infectious rates are so far not known. It is demonstrated the transmissibility of HGV by blood transfusion, hemoderivates and needle sharing but these sources of risk can't be responsible for the 10% prevalence on the general population. Mother to infant8 and close sexual13,14,15 contact transmission are currently thought to contribute most to the high prevalence seen worldwide. Though we haven't collected data concerning sexual activity of donors, in our series there was no association between HGV-RNA carrier state and more established markers of risky sexual behavior like HBsAg and anti-HIV (Table 1). However, a recently published cross-sectional study conducted with residents in the city of São Paulo detected an association between HGV and the number of sexual partners13. We haven't observed an association between HGV presence and elevation of ALT, corroborating that, to the healthy host, HGV causes no detectable hepatic damage.
An important issue in estimating an infectious agent prevalence by PCR is the choice of the primer system. We observed that primers spanning the 5'NCR detected more viremic donors than the NS3 nested-PCR, suggesting that HGV may be commonly present in a high viral load on the donor blood (Table 2) and analogously to HCV, the 5'NCR is conserved among isolates being the target of choice for PCR detection.
Among several sociodemographic variables studied, we observed a positive association with black race and a trend to higher positivity on donors reporting lower educational background. A significant association with black race has also been observed in South African blood donors4, although the factors for these racial differences are not known. It might be noted that we found an odds ratio of 4.5 of black/mulatto race over Caucasians, taking the latter as reference, but the prevalence on our Caucasian population is very high as well (7.5%), suggesting that this group is also exposed to the factor(s) responsible for the high prevalence of HGV-RNA in the general population. It should be pointed that this association remains statistically significant even after controlling for educational level (OR = 3.2, 95%CI 1.6;6.6), what was necessary since statistical analysis showed a trend for black/mulatto donors to have fewer years of education (c2 for trend p < 0.001).
It is known that viremic carriers represent only part of the exposed individuals, after the report that an antibody against the putative viral envelope (E2) protein correlates to virus clearance, i.e. absence of HGV-RNA16, and was shown to be present on a prevalence 2-3 times that of the PCR positive hosts. If this proportion proves to stand likewise to the population here studied, we could get to a 100% exposition rate on certain groups.
Phylogenetic analysis identified among Brazilian donors genotypes 1a, 1b, 2b and 3, according to the classification proposed by MUERHOFF et al.10. In contrast to reports from other populations19, "Brazilian" HGV isolates here described are not clustered, since they present higher sequence identity to isolates from USA, Japan, Germany and West Africa than to other Brazilian isolates, possibly reflecting the racial mixed composition of the Brazilian population. Alternatively, this may be due to the low number of isolates analyzed (nine). Isolate g87958 is placed on a separate branch of the tree (Fig. 2). In depth inspection of the alignment shows that this isolate probably represents a recombinant between genotype 1a and 2a since it harbors 5'NCR motifs from both genotypes. This can be due either by an amplification artifact or a genuine in vivo recombination, as observed on a Chinese isolate2.
Received: 09 October 2002
Accepted: 23 January 2003
- 1. ALTER, H.J.; NAKATSUJI, Y.; MELPOLDER, J. et al. - The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. New Engl. J. Med., 336: 747-754, 1997.
- 2. AN, P.; WEI, L.; WU, X. et al. - Evolutionary analysis of the 5'-terminal region of hepatitis G virus isolated from different regions in China. J. gen. Virol., 78: 2477-2482, 1997.
- 3. BASSIT, L.; KLETER, B.; RIBEIRO DOS SANTOS, G. et al. - Hepatitis G virus: prevalence and sequence analysis in blood donors of São Paulo. Vox Sang. (Basel), 74: 83-87, 1998.
- 4. CASTELING, A.; SONG, E.; SIM, J. et al - GB virus C prevalence in blood donors and high risk groups for parenterally transmitted agents from Gauteng, South Africa. J. med. Virol, 55: 103-108, 1998.
- 5. DAWSON, G.J.; SCHLAUDER, G.G.; PILOT-MATIAS, T.J. et al. - Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. J. med. Virol, 50: 97-103, 1996.
- 6. HOSMER Jr., D.M. & LEMESHOW, S. - Applied logistic regression New York. John Wiley, 1989.
- 7. KLEINMAN, S. - Hepatitis G virus biology, epidemiology, and clinical manifestations: implications for blood safety. Transfus. Med. Rev., 15: 201-212, 2001.
- 8. LEFRÈRE, J.-J.; SENDER, A.; MERCIER, B. et al - High rate of GB virus type C/HGV transmission from mother to infant: possible implications for the prevalence of infection in blood donors. Transfusion, 40: 602-607, 2000.
- 9. MUERHOFF, A.S.; SIMONS, J.N.; ERKER, J.C.; DESAI, S.M. & MUSHAHWAR, I.K. - Identification of conserved nucleotide sequences within the gb virus c 5'-untranslated region: design of PCR primers for detection of viral RNA. J. virol. Meth., 62: 55-62, 1996.
- 10. MUERHOFF, A.S.; SIMONS, J.N.; LEARY, T.P. et al. - Sequence heterogeneity within the 5'-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J. Hepat., 25: 379-384, 1996.
- 11. NAITO, H.; WIN, K.M. & ABE, K. - Identification of a novel genotype of hepatitis G virus in Southeast Asia. J. clin. Microbiol., 37: 1217-1220, 1999.
- 12. PICKERING, J.M.; THOMAS, H.C. & KARAYIANNIS, P. - Genetic diversity between hepatitis G virus isolates: analysis of nucleotide variation in the NS-3 and putative "core" peptide genes. J. gen. Virol., 78: 53-60, 1997.
- 13. SANTOS, G.R.; NISHIYA, A.S.; NASCIMENTO, C.M.R. et al. - Prevalence of GB virus C (Hepatitis G virus) and risk factors for infection in São Paulo, Brazil. Europ. J. clin. Microbiol. infect. Dis., 21: 438-443, 2002.
- 14. SCALLAN, M.F.; CLUTTERBUCK, D.; JARVIS, L.M.; SCOTT, G.R. & SIMMONDS, P. - Sexual transmission of GB virus C/hepatitis G virus. J. med. Virol., 55: 203-208, 1998.
- 15. STARK, K.; BIENZLE, U.; HESS, G. et al. - Detection of the hepatitis G virus genome among injecting drug users, homosexual and bisexual men, and blood donors. J. infect. Dis.,174: 1320-1323, 1996.
- 16. TACKE, M.; KIYOSAWA, K.; STARK, K. et al - Detection of antibodies to a putative hepatitis G virus envelope protein. Lancet, 349: 318-320, 1997.
- 17. THOMPSON, J.D.; HIGGINS, D.G. & GIBSON, T.J. - CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22: 4673-4680, 1994.
- 18. TUCKER, T.J. & SMUTS, H.E. - GBV-C/HGV genotypes: proposed nomenclature for genotypes 1-5. J. med. Virol., 62: 82-83, 2000.
- 19. VIAZOV, S.; RIFFELMANN, M.; KHOUDYAKOV, Y. et al. - Genetic heterogeneity of hepatitis G virus isolates from different parts of the world. J. gen. Virol., 78: 577-581, 1997.
Publication Dates
-
Publication in this collection
12 Dec 2006 -
Date of issue
Apr 2003
History
-
Received
09 Oct 2002 -
Accepted
23 Jan 2003