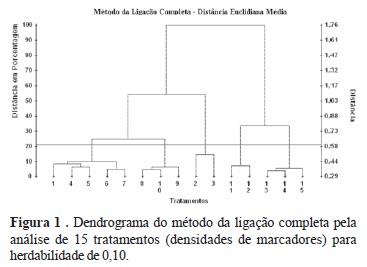
Simulation has contributed to the advancement of genomics in the different areas of genetic improvement. Genetic mappings were simulated using different densities of genetic markers to estimate phenotypic values of quantitative traits with heritabilities of 0.10; 0.40 and 0.70 in marker assisted selection (MAS). Cluster analysis with phenotypic performances was carried out to generate classification structures among the densities aiming to optimize QTL detection . The genetic simulation system (Genesys) was used to simulate three genomes (each consisting of a single characteristic differing in the heritability value) and the base and original populations. Each initial population was subjected to selection assisted by markers for 20 consecutive generations, in which selected parents mated selectively, between best and worst. The mapping using medium to high marker density showed efficiency in the phenotypic progress obtained with MAS. Smaller marker quantities are required to maintain power of QTL detection with increase in heritability. The cluster analysis indicated optimization and correspondence in phenotypic increases, when allowing the densities of 4 and 6 cM, 4, 6, 8 and 10 cM, and 6 and 8 cM for the heritabilities of 0.10; 0.40 and 0.70, respectively.
Cluster analysis; heritability; genome selection; simulation; QTL