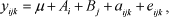This study aimed to evaluate the effects of major genes and population size on variance components estimation using four different types of selected populations. Variance components were estimated by classical and Bayesian methodologies, with three a priori information levels. In general, results from REML and Bayesian analyses with flat priors were similar. Except for Bayesian analysis with an informative prior, additive genetic variance estimates were not accurate in populations in which the trait is controlled by major genes. The use of pedigree information and records of all individuals back to the base-population was necessary to improve accuracy of variance component estimates, except for large populations in which the trait is controlled by a large number of genes.
computational simulation; Gibbs sampling; infinitesimal model; mixed models; REML; selection bias