Abstract
The purpose of this research was to elucidate the genetic control of orange corona color in carioca common beans (Phaseolus vulgaris). We made four crosses between carioca group cultivars that differed in respect to the presence or absence of an orange hilum corona color. The F2, F3, F1BC11, F1BC21, F2BC11 and F2BC21 phenotypic segregations were evaluated with a chi-square test which fitted with the hypothesis that one gene with a dominant allele is responsible for the orange corona color. All generations resulting from the four different crosses showed segregation patterns which agreed with the expected proportions. Our results show that the dominant G allele controls orange corona color in the carioca bean group.
carioca bean group; corona color; G locus; genetic inheritance
PLANT GENETICS
RESEARCH ARTICLE
Genetic control of orange hilum corona of carioca beans (Phaseolus vulgaris)
Juarez Pires TomazI; Vânia Moda-CirinoII; Nelson da Silva Fonseca JuniorII; Paulo Maurício RuasI
IDepartamento de Biologia Geral, Centro de Ciências Biológicas, Universidade Estadual de Londrina, Londrina, PR, Brazil
IIÁrea de Melhoramento e Genética Vegetal, Instituto Agronômico do Paraná, Londrina, PR, Brazil
Send correspondence to Send correspondence to: Vânia Moda-Cirino Instituto Agronômico do Paraná Área de Melhoramento e Genética Vegetal Caixa Postal 481, 86001-970 Londrina, PR, Brazil E-mail: vamoci@ iapar.br
ABSTRACT
The purpose of this research was to elucidate the genetic control of orange corona color in carioca common beans (Phaseolus vulgaris). We made four crosses between carioca group cultivars that differed in respect to the presence or absence of an orange hilum corona color. The F2, F3, F1BC11, F1BC21, F2BC11 and F2BC21 phenotypic segregations were evaluated with a chi-square test which fitted with the hypothesis that one gene with a dominant allele is responsible for the orange corona color. All generations resulting from the four different crosses showed segregation patterns which agreed with the expected proportions. Our results show that the dominant G allele controls orange corona color in the carioca bean group.
Key words: carioca bean group, corona color, G locus, genetic inheritance.
Introduction
The bean market is influenced by bean characteristics such as coat color, size and shape, cooking time, texture, flavor, taste and bean broth color. Studies on the genetic control of the testa color of beans have been carried out for more than seventy years, and a review of the genes studied until now had been published by Basset (2004).
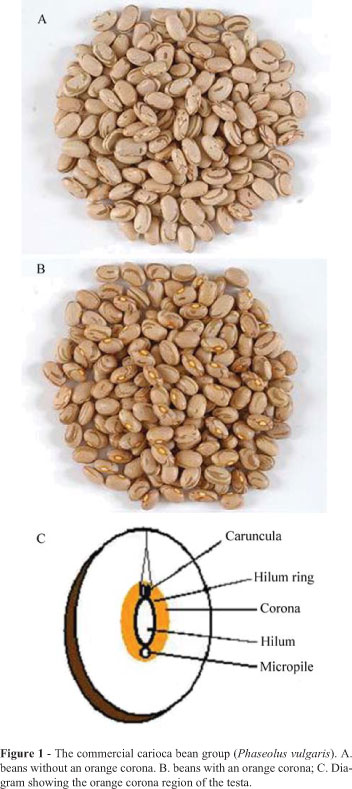
Brazil is the most important producer of beans and has an annual production of around three millions tonne (FAO, 2006), the most widely cultivated beans belonging to the carioca bean (Phaseolus vulgaris) commercial bean group. Carioca beans generally have a cream-colored testa, or seed coat, with clear brown stripes, but some have an orange hilum corona (Figure 1), which has been erroneously correlated with low grain cooking quality so that beans with this type of corona have a low market price and farmers avoid cultivating seeds with this trait. Disagreement occurs in regard to the genetic mechanism responsible for the presence of a corona, however the number of genes involved in the inheritance of this trait is lower than the genes involved in the genetic control of seed coat color (Barigozzi and Conti, 1981; Bassett, 1995; Mendonça et al., 1998; Bassett, 2003). A knowledge of the genetic control mechanisms responsible for the presence of an orange corona in carioca beans is important for the development of breeding programs for the development of carioca cultivars with high commercial acceptance.
The presence of a hilum corona in beans is controlled by the Cor locus, but the color of the corona depends of the expression of other genes (Leakey, 1988). The phenotypic segregation observed in the F2 offspring of some P. vulgaris crosses (P45 x Embrapa 201-Ouro, P45 x Carioca 300V and Carioca 300V x Embrapa 201-Ouro) has shown that at least two genes control the corona color trait, the Cor locus being responsible for the presence of a corona and the corona color being controlled by the interaction of three other loci (B, D, and G), the dark corona having the BBDDCorCorgg genotype and the yellow corona the BbddCorCorGG genotype (Mendonça et al., 1998). Basset (1995) suggested that beans with a dark hilum corona resulted from the pleiotropic effect of Vlae alleles, which are also responsible for pink flowers, so that plants with pink flowers would produce beans with dark corona while plants with white flowers would produce beans with no corona. Basset (1995) also suggested that another possible explanation for the genetic control of this trait could be strong genetic linkage between the genes for dark corona and pink flowers. However, the data obtained by Mendonça et al. (1998) disagree with this conclusion because the genotypes studied by these authors showed plants with white flowers and dark corona, which did not agree with a pleiotropic effect of the Vlae gene or a genetic linkage between the loci for flower color and dark hilum corona color.
The purpose of our research was to clarify the genetic control of orange hilum corona color in the commercial carioca bean group and assess the utility of this information for the genetic improvement of carioca-type beans.
Material and Methods
Plant material and crossing method
We investigated commercial carioca-group bean cultivars (Phaseolus vulgaris) contrasting in respect to the presence of an orange corona around the hilum ring (Figure 1), the cultivars being the orange corona cultivars Aporé, IAPAR-14 and IAPAR-57 which were used as female parents (P1) and the Carioca, FT-Paulistinha and Rudá cultivars which had no orange corona and were used as male parents (P2). Seed material for these plants was obtained from bean germplasm bank of the Paraná State Agronomic Institute (Instituto Agronômico do Paraná, IAPAR) and all the generations of the plants were grown in the greenhouse of the IAPAR, Londrina, PR, Brazil. The crosses were made according to Bliss (1980) and we evaluated the progenies of the crosses IAPAR-14 x Carioca, IAPAR-14 x FT-Paulistinha, IAPAR-57 x Carioca and Aporé x Rudá.
The presence or absence of an orange hilum corona is determined prior to fertilization (Ramalho et al., 2000) because the testa develops from the maternal ovule tegument (Esau, 1966) and its phenotype is thus determined by the genetic constitution of the female parent plant. We evaluated the phenotypic segregation of the orange corona trait in the P1, P2, F1, F2 and F3 generations and the backcross generations F1BC11, F1BC21, F2BC11 and F2BC21. The F1 seeds were used as female parents to get the BC11 and BC21 backcrosses. A random sample of 300 F1 beans and 90 BC1 beans were sown, then a random sample of 150 plants from each F1 population and a random sample of 15 plants from each BC11 and BC21 populations were selected and their F2, F1BC11 and F1BC21 progenies evaluated for the presence of orange hilum corona. To obtain the F3, F2BC11 and F2BC21 progenies we planted samples of 50 beans from each individual F2, F1BC11 and F1BC21 plant and when the plants were at the physiological maturation stage one pod from each F1BC11, F1BC12, F2, F3, F2BC11 and F2BC21 plant was evaluated for the presence of orange corona.
Statistical analysis
A chi-square test (Steel et al., 1997) was used to verify if the F2, F3, F1BC11, F1BC21, F2BC11 and F2BC21 phenotypic segregations fitted the hypothesis that one gene with a dominant allele controls the orange hilum corona trait. The Genes software (Cruz, 2001) was used to calculate the chi-square tests. The chi-square results were subjected to an homogeneity test (Strickberger, 1968) to check whether or not each generation was uniform and verify that the results could only be explained by the dominant monogenic inheritance genetic hypothesis.
Results and Discussion
All the F1 plants showed an orange hilum corona (Figure 1), while in the F2 generation the proportion of orange corona to non-orange corona segregates was 3:1 and in the F3 generation the segregant ratio was 5:3 for orange corona to non-orange corona segregates (Table 1). In the backcrosses the segregation ratio for orange corona to non-orange corona segregates was 1:0 for the F1BC11 generation, 1:1 for the F1BC21 generation, 7:1 for the F2BC11 generation and 3:5 for the F2BC21 generation (Table 1). These results indicate that a single gene with a dominant allele controls the orange hilum corona trait in the carioca bean group. In the four different crosses (IAPAR-14 x Carioca, IAPAR-14 x FT-Paulistinha, IAPAR-57 x Carioca, and Aporé x Rudá) all the evaluated progenies (F1 F2, F3, F1BC11, F1BC21, F2BC11 and F2BC21) showed segregation ratios supporting the hypothesis of a dominant allele controlling the presence of orange corona trait (Table 1). The chi-square homogeneity test for each generation demonstrated that the data obtained for each cross grouped in only one population (Table 2).
As discussed in the introduction, the hilum corona trait is controlled by the Cor locus and the G locus determines the presence of yellow-brownish pigments in the seed coat and its expression may be epistatic in relation to the C or J loci responsible for the color of the testa (Leakey, 1988), Mendonça et al. (1998) having shown that at least two independent genes are responsible for the presence of the orange corona trait and that in addition to the Cor locus three other loci (B, D, and G) must interact in the expression of corona color. Mendonça et al. (1998) also reported that interaction between the B and G loci is important for the expression of orange corona color in the carioca bean group, which presents the recessive allele b in the homozygote. Our data shows that only the G and Cor loci are needed for the expression of orange corona color in the commercial carioca bean group. This agrees with the findings of Basset et al. (2002), who reported that the orange corona phenotype occurs in Wagenaar and Enola common bean plants with the G-bb genotype.
In our study, the cultivars used as parents belonged to the carioca group and differed regarding the presence or absence of an orange hilum corona. In fact, all the plants analyzed by us had the same genotype for the corona trait (bbddCorCor), differing only regarding the presence of the G allele. Thus, the genotype of the parents with an orange corona (IAPAR-14, IAPAR-57, and Aporé) is bbdddCorCorGG and the genotype of the parents without the orange corona trait (Carioca, FT-Paulistinha, and Rudá) is bbddCorCorgg.
Our conclusion that the presence of an orange corona color in the carioca bean group is controlled by only one gene with a dominant allele is an important contribution for future breeding programs of this commercial bean group. The orange corona trait, wrongly supposed to correlate to low grain-cooking quality, is an undesirable characteristic because it is used by grain traders to justify a lower price for beans with this characteristic. Our results show that backcrossing can easily eliminate this trait from cultivars with otherwise good agronomic performance. The genotypes of desirable plants can be identified using progeny testing. In conclusion, the presence of orange corona trait in commercial carioca bean group may be controlled by only two genes, the Cor locus responsible for the presence of the hilum corona trait and the G locus for the orange corona color. These data are important for future breeding programs of carioca bean group market classes.
Acknowledgments
The authors gratefully acknowledge support by the Brazilian agencies Agronomic Institute of Paraná (Instituto Agronômico do Paraná, IAPAR) and the Brazilian National Counsel of Technological and Scientific Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq).
Internet Resources
Food and Agriculture Organization of the United Nations (2006) FAO Yearbook. http://www.fao.org. Accessed in July 3, 2006.
Received: August 24, 2006; Accepted: April 2, 2007.
Senior Editor: Ernesto Paterniani
- Barigozzi C and Conti L (1981) Heredity of seed coat color in Phaseolus vulgaris: Variegation and limitation of the coloured field. Genetica Agraria 35:209-217.
- Bassett MJ (1995) The dark corona character in seedcoat of common bean co-segregates with the pink flower allele vlae J Am Soc Hort Sci 120:520-522.
- Basset MJ (2003) Inheritance of yellow corona and hilum ring in seed coat of mayacoba market class common beans with genotype P [Cr} gy J g b Vlae Rk. J Am Soc Hort Sci 128:721-723.
- Basset MJ (2004) List of genes - Phaseolus vulgaris L. Ann Rev Bean Improv Coop 47:1-24.
- Basset MJ, Lee R, Otto C and McClean PE (2002) Classical and molecular genetic studies of the strong greenish yellow seed coat color in Wagenaar and Enola common bean. J Am Soc Hort Sci 127:50-55.
- Bliss FA (1980) Common bean. In: Fehr WR and Hadley HH (eds) Hybridization of Crop Plants. American Society of Agronomy - Crop Science Society of America, Madison, pp 273-284.
- Cruz CD (2001) Programa GENES: Versão Windows; Aplicativo Computacional em Genética e Estatística. Ed. UFV. Viçosa, 648 pp.
- Esaú K (1966) Anatomy of Seed Plants. 5th edition. John Willey &Sons, Inc., New York, 376 pp.
- Leakey CLA (1988) Genotypic and phenotypic markers in common bean. In: Gepts P (ed) Genetic Resources of Phaseolus Beans. Kluwer Academic Press, Boston, pp 245-327.
- Mendonça HA, Santos JB, Ramalho MAP and Ferreira DF (1998) Genetic control of the fungus Colletotrichum lindemuthianum (Sacc. & Magn.) Scrib reaction and corona color in the common bean (Phaseolus vulgaris L.). Genet Mol Biol 21:335-341.
- Ramalho MAP, Santos JB and Pinto CABP (2000) Genética na Agropecuária. 2 edição. Ed. UFLA, Lavras, 472 pp.
- Steel RGD, Torrie JH and Dickei, DA (1997) Principles of Procedures of Statistics a Biometrical Approach. 3rd edition. Mc Grow Hill, Boston, 666 pp.
- Strickberger MW (1968) Genetics. The Macmillan Company, New York, 868 pp.
Send correspondence to:
Publication Dates
-
Publication in this collection
30 Aug 2007 -
Date of issue
2007
History
-
Accepted
02 Apr 2007 -
Received
24 Aug 2006