Abstract
The availability of a useful tool for simple and timely detection of the most important virulent varieties of Escherichia coli is indispensable. To this end, bacterial DNA pools which had previously been categorized were obtained from isolated colonies as well as selected in terms of utilized phenotype; the pools were assessed by two PCR Multiplex for the detection of virulent E. coli eaeA, bfpA, stx1, stx2, ipaH, ST, LT, and aatA genes, with the 16S gene used as DNA control. The system was validated with 66 fecal samples and 44 wastewater samples. At least one positive isolate was detected by a virulent gene among the 20 that were screened. The analysis of fecal samples from children younger than 6 years of age detected frequencies of 25% LT positive strains, 8.3% eae, 8.3% bfpA, 16.7% ipaH, as well as 12.5 % aatA and ST. On the other hand, wastewater samples revealed frequencies of 25.7% eaeA positive, 30.3% stx1, 15.1% LT and 19.7% aatA. This study is an initial step toward carrying out epidemiological field research that will reveal the presence of these bacterial varieties.
virulence genes; wastewater; fecal; screening system; Escherichia coli
MEDICAL MICROBIOLOGY
A DNA pooling based system to detect Escherichia coli virulence factors in fecal and wastewater samples
LuzMaría Chacón JI, * * Corresponding Author. Mailing address: Sección Infección-Nutrición, Instituto de Investigaciones en Salud (INISA), University of Costa Rica, Montes de Oca, Costa Rica.; E-mail: luz.chacon@ucr.ac.cr ; Lizeth Taylor CII; Carmen Valiente AIII; Irene Alvarado PII; Ximena Cortés BII
ISección Infección-Nutrición, Instituto de Investigaciones en Salud (INISA), University of Costa Rica, Montes de Oca, Costa Rica
IIVirology Division, Microbiology Department, University of Costa Rica, Montes de Oca, Costa Rica
IIILaboratorio Nacional de Aguas, Instituto Costarricense de Acueductos y Alcantarillados (AyA), La Unión, Costa Rica
ABSTRACT
The availability of a useful tool for simple and timely detection of the most important virulent varieties of Escherichia coli is indispensable. To this end, bacterial DNA pools which had previously been categorized were obtained from isolated colonies as well as selected in terms of utilized phenotype; the pools were assessed by two PCR Multiplex for the detection of virulent E. coli eaeA, bfpA, stx1, stx2, ipaH, ST, LT, and aatA genes, with the 16S gene used as DNA control. The system was validated with 66 fecal samples and 44 wastewater samples. At least one positive isolate was detected by a virulent gene among the 20 that were screened. The analysis of fecal samples from children younger than 6 years of age detected frequencies of 25% LT positive strains, 8.3% eae, 8.3% bfpA, 16.7% ipaH, as well as 12.5 % aatA and ST. On the other hand, wastewater samples revealed frequencies of 25.7% eaeA positive, 30.3% stx1, 15.1% LT and 19.7% aatA. This study is an initial step toward carrying out epidemiological field research that will reveal the presence of these bacterial varieties.
Key words: virulence genes, wastewater, fecal, screening system, Escherichia coli
INTRODUCTION
Diarrhea continues to be a major cause of illness and death worldwide. According to the World Health Organization (WHO), 1.4 million pre-school children die annually due to diarrhea resulting from intestinal infection (19).
The Pan-American Health Organization (PAHO) reports that the average rate of death from acute diarrhea in children younger than 5 years of age in Latin America and the Caribbean is 4.1%. The regions with the highest rates are Central America (7.8%), the Caribbean (5.5%), and 5.1% in the Andean region (23). Costa Rica, for example, recorded 450 deaths due to diarrhea during 2001-2005, of which only 20 were found to have an identified causal agent (18).
Escherichia coli is a Gram negative bacillus, a normal microorganism in humans, but one which can produce symptoms ofdiarrhea when virulent factors are acquired, among which are enterotoxins, adhesines, and colonization factors (33).
Diarrheogenic varieties of E. coli exhibit a wide range of clinical symptoms that include traveler's diarrhea and hemorrhagic diarrhea (24) as well as cases produced by zoonosis (14).To date, six varieties have been clearly described and characterized by virulence factors capable of causing diarrhea in humans: 1) enterotoxigenic E. coli or ETEC, whose most distinctive genes are the stable thermotoxin -st and thermolabile -lt -; 2) enteropathogenic E. coli or EPEC, whose characteristic genes are the intimine - eaeA and the bundle forming pilus - bfp -); 3) shiga-toxin E. coli or STEC, whose toxins are encoded in the stx1 y stx2 genes, 4) enteroinvasive E. coli or EIEC, one of whose characteristic virulence traits is the ipaH, which belongs to the invasion plasmid; 5) enteroaggregativeE. coli or EAEC, with the pCVD432 plasmid, for which the aatA gene is one of the most stable regions; and 6) diffusely adherent E. coli or DAEC, whose virulence genes have yet to be fully profiled (11, 22).
A number of different procedures have been described for identifying these pathogens since this cannot be carried out based on their phenotypical features. These strategies have ranged from cultivating cells to bio-molecular identification, such as Multiplex PCR (1, 3, 11, 20, 21).This means that routine detection of these varieties is costly. Moreover, the literature demonstrates that the frequency of virulent E. coli varieties is much lower than that of non-virulent varieties, thus making it necessary to screen various isolates per sample in order to detect them (10).Costa Rica and most other Latin American countries lack epidemiological data on the circulation of diarrheogenic E. coli as well as on its impact on public health. The main goal of the present study was to design a quick and easy screening system that would make analysis of the greatest number of E. coli isolates possible, as well as to determine the applicability of the system in both fecal samples from children less than 6 years of age and in wastewater samples taken from stabilization or settling ponds.
METHODOLOGY
Samples
Sixty six fecal samples were collected through a community project at the University of Costa Rica (TCU-350) with children younger than 6 years of age in Montes de Oca county in San José province, Costa Rica, during February and March of 2007. These samples were inoculated in MacConkey agar (Oxoid®) within 12 hours after collection, and were incubated at 35 ºC for 24 hours. Moreover, 24 water samples were collected for over a year (from September of 2007 until December of 2008) from inlet and outfall points of the Costa Rican Water Supply and Sewer Institute´s settling ponds located in Nicoya, Cañas, Liberia, Santa Cruz, and Pérez Zeledón. The wastewater samples were processed by the most probable number (MPN) technique for fecal coliforms as described by the American Public Health Association (2). After the presumptive phase, a mix was made of all tubes that tested positive by dilution. Each subsample (subsamples being understood as each dilution of an analyzed sample that had tested positive) was then inoculated into MacConkey agar (Oxoid®) and incubated at 35 ºC for 24 hours.
Gene pool system
Twenty colonies from the bacterial cultures of the feces and wastewater samples obtained in MacConkey agar (Oxoid®) were inoculated in a single tube of soy trypticase broth (Oxoid®) and then incubated at 35 ºC for 24 hours. In order to include atypical E. coli morphotypes, all the pools were prepared with 25% lactose negative colonies and 75% lactose positive colonies.To be able to assess the detection limit of the method, bacterial pools containing positive control colonies and strains with no virulence factors were utilized; the ratio used was 1:19 (1 positive colony was inoculated with a known virulence factor, along with 19 bacterial strains with not known virulence factors). Pools with 1+1:18 ratios, in which colonies that were carriers of different virulence factors were mixed with strains with not known virulence factors, were also evaluated.
DNA extraction and quantification
The extraction of the DNA was carried out in accordance with the Franck thermic protocol (7). The extracted DNA material was stored at -70 ºC until being used. The quantification of the DNA from each pool was carried out with an Epependorff Plus® biophotometer, with a dilution factor of 1:200.
Polymerase Chain Reaction (PCR)
Two Multiplex PCR and one simple PCR, which had been standardized in the laboratory, were used to process the gene pool samples. For the PCR Multiplex, the following reagent concentration was utilized: free buffer Mg 1X Fermentas®, MgCl2 1.75 mM Fermentas®, dNTP 200 µM Fermentas®, 0.6 U of Taq polymerase Fermentas® per reaction, and 4 µl of bacterial DNA. The primers utilized for the PCR Multiplex 1 were as follows: 5´GAC CCG GCA CAA GCA TAA GC 3´ and 5´CCA CCT GCA GCA ACA AGA GG 3' for the eaeA gene (384 pb, 0.04 µg/reaction), 5´AAT GGT GCT TGC GCT TGC TGC 3´ and 5´ GCC GCT TTA TCC AAC CTG GTA3´ for the bfpA gene (324 pb, 0.03 µg/reaction), 5´CTG GATTTA ATG TCG CAT AGT G3´ and 5´AGA ACG CCC ACT GAG ATC ATC3´ for the stx1 gene (150 pb, 0.045 µg/reaction), 5´GGC ACT GTC TGA AAC TGC TCC 3´ and 5´TCG CCA GTT ATC TGA CAT TCT G3´ for the stx2 gene (250 pb, 0.06 µg/reaction)(13) and 5'GTTCCTTGACCGCCTTTCCGATA CCGTC3' and 5'GCCGGTCAGCCACCCTCTGAGAGTAC 3' for the ipaH gene (600 pb,0.05 µg/reaction) (4). The primers utilized for the Multiplex PCR 2 were as follows: 5´ TCT GTA TTG TCT TTT TCA CC 3´ and 5´ TTA ATA GCA CCC GGT ACA AGC 3´ for the st toxin (186 pb, 0.4 µg/reaction) (8), 5' TCT CTA TGT GCA TAC GGA GC 3' and 5' CCA TAC TGA TTG CCG CAA T 3' for the lt toxin (322 pb, 0.02 µg/reaction) (30) and 5´CTG GCG AAA GAC TGT ATC AT 3´ and 5´CAATGTATAGAAATCCGCTGTT 3´ for a segment of the pCVD423 aatA gene (630 pb, 0.045 µg/reaction) (27). Finally, a simple PCR with MasterMix Fermentas® 2X was used for the 16S; as primers, 5'GGGATGAAAGTTAATACCTTTGCTC 3' and 5' TTCCCGAAGGCACATTCT 3' (584 pb) were used (31). The amplification was done in a DNA thermal cycler (Applied BioSystem®) using 5 min at 95 °C for initial denaturation followed by 35 cycles of 92°C (denaturation), 55 °C (annealing) and 72 °C (primer extension), for 45 seconds each, with a final 10 min extension at 72°C). For the simple PCR, the amplification conditions were a initial denaturation for 5 minutes at 94 ºC, followed by 35 cycles of PCR divided in 94 ºC (denaturation), 56 ºC (annealing) and 72 ºC (primer extension), for 30 seconds each, with a final extension of 2 minutes at 72 ºC. The detection limit for all the PCR Multiplex and simple PCR reactions was assessed byutilizing quantified bacterial DNA dilutions (between 10 and 60 ng/µl) and the specificity of the PCR was assessed by using other members of the family Enterobacteriaceae. PCR products were visualized by 2% agarose gels (TopVisionTM Fermentas®) with 0.05% ethydium bromide. The electrophoresis processes were carried out in a Fisher Scientific® electrophoresis chamber at 13.5 V/cm for 60 minutes. DNA fragment sizes were compared to a molecular weight marker (Fermentas®), with a 50 base pair ladder.
RESULTS
PCR
The optimum conditions of each PCR reaction were standardized in the laboratory and detailed in the methodology.The minimum concentration of DNA detected was of 20 µg/ml for the 16S, the stx1, stx2, and ipaH genes, and 30 µg/ml for the eaeA and bfpA genes.In the Multiplex 2 reaction, the minimum concentration of DNA detected was 40 µg/ml. For which extractions of DNA with concentrations of between 60 and 200 µg/ml were used.In the specificity assays amplification was observed only for the 16S gene of Shigella sp. strain, whereas no amplification was observed in any of the other genes and strains.
Gene pool system
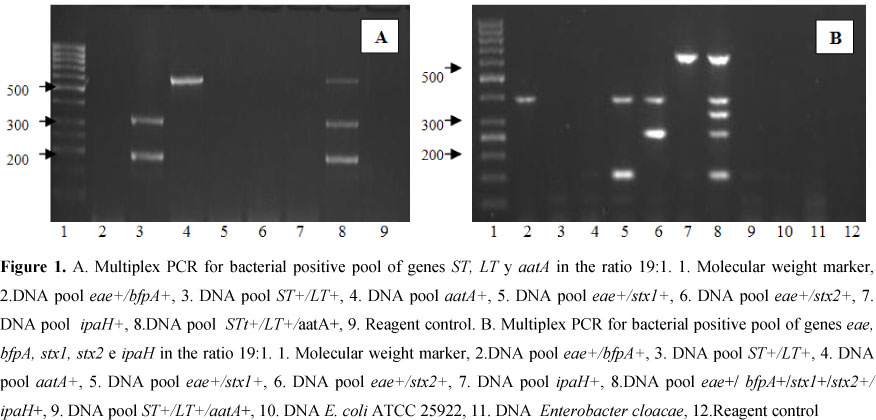
The detection limit of the gene pool method was evaluated in order to determine the capacity of detecting the virulence genes of pathogenic E. coli strains, which were present in the pool in a much lower concentration as compared to the predominant non virulent E. coli strains (1+19). Before the PCR analysis, the DNA obtained for each gene pool was quantified, and positive detections were obtain from the DNA pools in which only one strain was the carrier of virulence factors (1:19); this was done in quintuplicate for each gene under study (Figure 1). To evaluate the simultaneous detection of the genes, mixed pools were prepared from the positive control strains, an effective detection was achieved for each one of the specific genes. This was carried out in quintuplicate for the eaeA+/bfpA+, stx1, stx2, ipaH combinations, and for the ST+/LT+, aatA combination.
Fecal samples
Of the 66 samples analyzed, 1 was discarded since the 16S gene was not detected by PCR; samples showing negative results for the 16S gene were excluded since with the established conditions, their detection would be found to be below the sensitivity limit. Thirty-seven percent of the samples yielded negative results, 34% of the samples tested positive for more than one virulence factor, and 24% of the samples tested positive for just one virulence factor. The specific results are detailed in Table 1.
Wastewater samples
Of the 24 samples taken from settling ponds, which were analyzed as 77 subsamples, no samples were discarded since in every case at least one subsample showed amplification of the 16S gene. Thirteen samples were taken from pond inlets, and 11 were taken from their out fall points. Of the total of samples analyzed, 46% were negative for all virulence factors under study, 33.3% were positive for at least one virulence factor, and 12.5% were positive for just one virulence factor. The relative frequencies found are detailed in Table 1.
DISCUSSION
The gene pool system is an innovative element in this study. It has been widely documented that detection of diarrheagenic varieties of E.coli is methodologically complex because this bacterial group is found in low concentrations in fecal samples, and then as a result, analysis from 1 to 5 isolates can yield low detection frequencies (10, 16). We propose that this newly described tool can be used to overcome this limitation.
To work adequately, the gene pool system requires adjustments and standardization of the quantity of genetic material obtained in order to generate reproducible results (28). In the case of this PCR, to standardize the optimum conditions for amplification and to prevent false negatives because of low DNA concentration, assays were carried out to determine the minimum concentration of DNA necessary for detection of the expected products (between 60 and 200 µg/ml); thus, the results obtained are reproducible.
Moreover, the implemented detection system was assayed in two distinct matrixes in order to evaluate its versatility. Fecal samples were chosen due to the fact that in Latin America it is common to find these bacterial groups among, both symptomatic and asymptomatic populations (26). On the other hand, settling ponds were selected because they are ecosystems with a great variety of microorganisms where it is likely to find potentially virulent strains (32).
The feasibility of detecting diarrheagenic E. coli in the analysis of samples from wastewater in settling ponds by enhancing the most probable number (MPN) approach is evident. This is what makes this procedure much more accessible than current alternatives, such as concentration techniques by "in situ" filters, or filtering through membranes (2).
An important thing to note is that all the positive dilutions have to be analyzed by the enhanced approach since, the presence of microorganisms does not follow a statistically normal distribution (2). This fact was corroborated in this study upon finding the presence of microorganisms in different dilutions of a given sample, regardless of the composition of dilutions themselves.
The settling ponds studied have retention periods of approximately three months, and their effectiveness, from the microbiological standpoint, is measured by the capacity to diminish the microbial load in at least three logarithms (25). Nevertheless, the survival of pathogenic microorganisms has not been studied in detail. In addition to the detection of the genes under analysis, differences were found among the profiles of genes across the samples taken from the pond inlet and outfall points (Table 1).
This phenomenon can be explained from the theoretical point of view given that the genome of diarrheagenic varieties of E. coli is common to varieties found in the wild, which allows both groups of bacteria to survive in the greater environment (9). Moreover, there is evidence that some varieties, such as the Shiga toxin producing E. coli, can withstand adverse conditions such as changes in temperature or pH level, and even desiccation. Such evidence suggests that settling ponds are ecosystems with the ability to promote survival of potentially pathogenic bacteria even though the ponds are efficient in reducing the number of microorganisms that end up exiting the system. This survival may mean that the ponds recirculate the major virulent varieties of E. coli and release them into the environment at different moments, which could explain the variation in the profiles of virulent genes found across the ponds inlet and outfall points.
On the other hand, the system proved capable of detecting diarrheagenic E. coli in fecal samples as well. It is important to emphasize that although that the samples were taken from asymptomatic children, the frequencies detected were greater than those usually described, in which only 1 to 3 isolations per sample are analyzed (6). Among the hypotheses to explain this finding are the following: improved detection thanks to the use of gene pools, a high prevalence of pathotypes in the population, and the fact that these pathogenic varieties are regulated by the surroundings (17, 9). However, future studies are needed in order to analyze this phenomenon in detail.
In the fecal and wastewater samples analyzed, genes were simultaneously found that are characteristic of both ETEC and EPEC; this is a trend that has been described for a number of years, and which at the clinical level produces more severe symptoms than those generated by either of these varieties individually (5). These findings can serve as a guide for better treatment in the health sector for patients with acute diarrhea that present these co-infections. Nonetheless, studies of this nature are scarce, as mentioned previously, due to routine analysis being carried out by means of profiling 1 to 3 isolations on an individual basis, which could overlook identification of other pathogenic varieties present in a given sample.
Another important finding is the high frequency at which the eaeA gene was detected, both in wastewater and fecal samples; this could indicate the presence of atypical ECEP varieties in the country, which is congruent with findings worldwide, where the atypical EPEC varieties are more frequent than typical ones (1).
Finally, although methodologies currently exist to analyze microorganisms directly in the sample which improves response time, with this system, the cultivation of microorganisms ensures their viability and avoids the problems created by substance inhibitors in the sample for detection by PCR. (12, 29)
To summarize, the present study demonstrates the efficiency of implementing a gene pool system for detecting diarrheagenic E. coli. This tool is useful for analysis of wastewater and fecal samples, and its implementation in the field is an initial step toward carrying out epidemiological studies to assess the incidence, prevalence, and persistence of certain diarrheagenic E. coli varieties. This information can make it possible to orient authorities as to the proper means of environmental control when handling wastewater, as well as to guide those in the clinical field as to the best course of treatment for patients infected with one or more pathogenic varieties of E. coli.
ACKNOWLEDGEMENTS
To the Unidad de Aguas Residuales del Laboratorio Nacional de Aguas, Instituto Costarricense de Acueductos y Alcantarillados (AyA). To Dra. Elizabeth Abrahams for donation of fecal samples collected by TCU-350 "Diagnóstico parasitológico en comunidades costarricenses" of Parasitology Section of Microbiology Faculty of the Costa Rica University. To Dra. Olga Torres of Instituto de Nutrición de Centro América y Panamá (INCAP) y Dra Mercedes Cáceres de la Universidad de León for donation control strains for this study. To Harlan Wolfe and Clyde McCoy from CDRC/University of Miami for kind suggestions to improve the paper and to the Scientific Publications Workshop, INISA, UCR/CDRC,UM, for advice and support.
Project 04-N-2003 "Characterization of enterotoxigenic Escherichia coli (ETEC) isolates from Guatemala and Costa Rican children" for Network for Research and Training in Tropical Diseases in Central America (NeTropica).
Project "Desarrollo de métodos moleculares y de cultivo celular para el estudio de factores de virulencia de E. coli patogénicas" for "Fondo de Incentivo para Reinserción de Investigadores de la Comisión Nacional para Investigaciones Científicas y Tecnológicas (CONICIT)"
Submitted: July 15, 2011
Returned to authors for corrections: December 22, 2011
Approved: June 07, 2012
- 1. Abe, C.M.; Trabulsi, L.R.; Blanco, J.; Blanco, M.; Dahbi, G.; Blanco, J.E.; Mora, A.; Franzolin, M.R.; Taddei, C.R. Martinez, M.B.; Piazza, Chacón-J., L.M. et al. DNA pooling based system to detect E. coli R.M.F.; Elias, W.P. (2009). Virulence features of atypical enteropathogenic Escherichia coli identified by the eae (+) EAF-negative stx (-) genetic profile. Diagn Microbiol Infect Disease 64 (4), 357-365.
- 2. American Public Health Association; American Water Works Association; Water Environment Federation. (2005). Standard Methods for the Examination of Water and Wastewater American Public Health Association, Washington, D.C.
- 3. Anbazjagan D, Seok Mui W, Mansor M, Siok Yan GO, Yasim Yusof M, Devi Sekaran S. (2011). Development of Conventional and Real-Time Multipex PCR Assays for the detection of nosocomial pathogens. Braz. J. Microbiol. ;42:448-458
- 4. Aranda K, Fagundes-Neto U, Scaletsky A. (2004). Evaluation of Multiplex PCRs for diagnosis of Infection with Diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol; 42 (12), 5849-5853
- 5. Crane, J.; Choudhari S, Naeher, T.; Duffey, M. (2006). Mutual enhacenment of virulence by Enterotoxigenic and Enteropathogenic Escherichia coli Infect Immun 74 (3), 1005-1515
- 6. Estrada-García, T.; Lopez-Saucedo, C.; Thomson-Bonilla, R.; Abonce, M..; Lopez-Hernandez, D.; Santos, J.I.; Rosado, J.L. (2009). Association of Diarrheagenic Escherichia coli pathotypes with infection and diarrhea among Mexican children and association of atypical enteropathogenic E. coli with acute diarrhea. J Clin Microbiol 47 (1), 93-98
- 7. Franck, S.; Bosworth, B.; Moon H.W. (1998). Multiplex PCR for Enterotoxigenic, Attaching and Effacing, and Shiga Toxin-Producing Escherichia coli Strains from Calves. J Clin Microbiol 36, 1795-1797.
- 8. Frankel, G.; Girón, J.A.; Valmassoi, J.; Schoolnik G.K. (1989). Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Mol Microbiol 3, 1724-1734.
- 9. Garmendia, J.; Frankel, G.; Crepin V. (2005). Enteropathogenic and Enterohemorrhagic Escherichia coli intections: Translocation, translocation, translocation. Infect Immun 73 (5), 2573-2585
- 10. Iijima, Y.; Tanaka, S.; Miki, K.; Kanamori, S.; Toyokawa, M.; Asari, S. (2007). Evaluation of colony-based examinations of diarrheagenic Escherichia coli in stool specimens: low probability of detection because of low concentrations, particularly during the early stage of gastroenteritis. Diagn Microbiol Infect Dis 58, 303-308
- 11. Karper, J.; Nataro, J.; Mobley H. (2004). Pathogenic Escherichia coli Nat Rev Microbiol 2, 123-14
- 12. Kong, R.Y.C.; Lee, S.K.Y.; Law, T.W.F; Wu, R.S.S. (2002). Rapid detection of six types of bacterial pathogens in marine waters buy multiplex PCR. Water Res. 36 (11), 2802-2812
- 13. López-Saucedo, C.; Cerna, J.F.; Villegas-Sepulveda, N.; Thompson, R.; Velazquez, F.R.; Torres, J.; Tarr, P.I.; Estrada-Gacía, T. (2006). Single Multiplex polymerase chain reaction to detect diverse loci associated diarrheagenic Escherichia coli Emerg Infect Dis 9 (1), 127-131
- 14. Loukiadis, E.; Kérourédan, M.; Beutin, L.; Oswald, E.; Brugère, H. (2006). Characterization of Shiga Toxin gene (stx) -positive and Intimin gene (eae) -positive Escherichia coli isolates from wastewater of slaughterhouses in France. Appl Environ Microbiol 72 (5), 3245-3251
- 15. Lui, Y.; Gilchrist, A.; Zhang, J.; Li, X. (2008). Detection of viable but nonculturable Escherichia coli O157:H7 bacteria in drinking water and river water. Appl Environ Microbiol 74 (5), 1502-1507
- 16. Manning, S.; Madera, R.; Schneider, W.; Dietrich, S.E.; Khalife, W.; Brown, W.; Whittan, T.S.; Somsel, P.; Rudrik, T. (2007). Surveillance for Shiga Toxin-producing Escherichia coli, Michigan, 2001-2005. Emerg Infect Dis 13 (2), 318-321
- 17. Mellies, J.; Barron, A.; Carmona, A. (2007). Enteropathogenic and Enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75 (9), 4199-4210
-
18Ministerio de Salud de Costa Rica y Organización Panamericana de la Salud. (2006) Mortalidad por enfermedades de declaración obligatoria en Costa Rica: Año 2005. Available at: http://www.ministeriodesalud.go.cr/mortalidad/mortalidad2005.pdf Accesed 5 August 2010
- 19. Moyo, S.J.; Maselle, S.Y.; Matee, M.I.; Langeland, N.; Mylvaganam, H. (2007). Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infectious Diseases 7 (92). Available at: http://www.biomedcentral.com/1471-2334/7/92
- 20. Müller, D.; Greune, L.; Heusipp, G.; Karch, H.; Fruth, A.; Tschäpe, H; Schmidt, M.A. (2007). Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73 (10), 3380-90.
- 21. Nataro, J.; Kaper, J.B. (1998). Diarrheagenic Escherichia coli Clin Microbiol Rev 11, 142-201.
- 22. Nishi, J.; Sheiikh, J.; Mizuguchi, K.; Luisi, B.; Burland, V.; Boutin, A.; Rose, D.J.; Blattner, F.; Nataro, J. (2003). The export of coat protein from Enteroaggregative Escherichia coli by specific ATP-binding cassette transporttes system. J Biol Chem 278 (46), 45680-45689
-
23Organización Panamericana de la Salud. (2009) Situación de Salud en las Américas: Indicadores Básicos 2009. Washington, D.C., Estados Unidos de América. Available at: http://new.paho.org/hq/dmdocuments/2009/IB_SPA_2009.pdf Accessed: 12 July 2011
- 24. Qadri, F.; Svennerholm, A.; Faruque, S.; Sack, B. (2005). Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18 (3), 465-483
- 25. Ramírez, J.; Valiente, C. (2010). Informe Anual 2009: Calidad de Aguas residuales en los sistemas de tratamiento operados y administrados por AyA y estudios especiales de interés institucional Instituto Costarricense de Acueductos y Alcantarillados, Laboratorio Nacional de Aguas, AyA.
- 26. Rúgeles, L.C.; Bai, J.; Martínez, A.J.; Vanegas, M.C.; Gómez-Duarte, O.G. (2010). Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int J Food Microbiol 138 (3), 282-286
- 27. Schmidt, H.; Knop, C.; Franke, S.; Alesksic, S.; Heesemann, J.; Karch, H. (1995). Development of PCR for screening of enteroaggregative Escherichia coli.J Clin Microbiol 33 (3), 701-705
- 28. Sham, P.; Bader, J.S.; Craig, I.; O'Donova, M.; Owen, M. (2002). DNA Pooling: a tool for large-scale association studies. Nat Rev Genet 3, 862-871
- 29. Stacy-Phipps, S.; Mecca, J.J.; Weiss, J. (1995). Multiplex PCR assay and simple preparation method for stool specimens detect Enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 33(5), 1054-1059
- 30. Toma, C.; Lu, Y.; Higa, N.; Nakasone, N.; Chinen, I.; Baschkier, A.; Rivas, M; Iwanaga, M. (2003). Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol 41 (6), 2669-2671
- 31. Tsen, H.; Lin, C.; Chi, W. (1998). Development and use of 16S rRNA gene targeted PCR primers for the identification of Escherichia coli cells in water. J Appl Microbiol 85, 554-560
- 32. Weintraub A. (2007). Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J Med Microbiol 56 (1), 4-8
- 33. Wolf-Rainer A. (2011). Megacities as source for pathogenic bacteria in rivers and their fate downstream. Int J Microbiol Available at: http://www.hindawi.com/journals/ijmb/2011/798292/
Publication Dates
-
Publication in this collection
19 Feb 2013 -
Date of issue
Dec 2012
History
-
Received
15 July 2011 -
Accepted
07 June 2012 -
Reviewed
22 Dec 2011