Abstract
In this study, oil degrading bacteria discovered from fish living near the oil ports at Karachi in Pakistan were characterized. The bacteria isolated from skin, gills, and gut in fish could consume crude oil as a source of carbon and energy. Total 36 isolates were tested using Nutrient Agar (NA) and MSA media with different crude oil concentrations (0.2%, 0.5%, 0.7%, 1%, 2%, and 5%) and 4 out of 36 isolates (two Gram positive and two Gram negative bacteria) were selected for further identification. 16S rRNA gene sequencing revealed that the isolates are related to Bacillus velezensis, Bacillus flexus, Pseudomonas brenneri and Pseudomonas azotoforman. Oil degrading potential of these bacteria was characterized by GC-MS analysis of degradation of oil components in crude oil as well as engine oil. We found that one (2, 6, 10, 14-Tetramethylpentadecane) out of 42 components in the crude oil was fully eliminated and the other oil components were reduced. In addition, 26 out of 42 oil components in the engine oil, were fully eliminated and the rest were amended. Taken together, these studies identify that B. velezensis, B. flexus, P. brenneri and P. azotoforman have high oil degrading potential, which may be useful for degradation of oil pollutants and other commercial applications.
Keywords:
oil degradation; bacteria; bioremediation; fish; Indian Ocean
Resumo
Neste estudo, bactérias degradadoras de óleo descobertas em peixes que vivem perto dos portos de petróleo em Karachi, no Paquistão, foram caracterizadas. As bactérias isoladas da pele, guelras e intestinos dos peixes podem consumir petróleo bruto como fonte de carbono e energia. No total, 36 isolados foram testados usando Agar Nutriente (NA) e meio MSA com diferentes concentrações de óleo bruto (0,2%, 0,5%, 0,7%, 1%, 2% e 5%) e 4 de 36 isolados (dois Gram positivos e duas bactérias Gram negativas) foram selecionadas para posterior identificação. O sequenciamento do gene 16S rRNA revelou que os isolados estão relacionados a Bacillus velezensis, Bacillus flexus, Pseudomonas brenneri e Pseudomonas azotoforman. O potencial de degradação do óleo dessas bactérias foi caracterizado pela análise de GC-MS da degradação dos componentes do óleo no óleo cru, bem como no óleo do motor. Descobrimos que um (2, 6, 10, 14-tetrametilpentadecano) de 42 componentes do óleo cru foi totalmente eliminado e os outros componentes do óleo foram reduzidos. Além disso, 26 dos 42 componentes do óleo do motor foram totalmente eliminados e o restante corrigido. Juntos, esses estudos identificam que B. velezensis, B. flexus, P. brenneri e P. azotoforman têm alto potencial de degradação de óleo, o que pode ser útil para a degradação de poluentes de óleo e outras aplicações comerciais.
Palavras-chave:
degradação de óleo; bactéria; biorremediação; peixe; Oceano Índico
1. Introduction
Pollution by oil discharges is a serious problem of the marine environment around the world that has adversely affect the biotic and abiotic processes (Lee et al., 2015LEE, K., BOUFADEL, M., CHEN, B., FOGHT, J., HODSON, P., SWANSON, S. and VENOSA, A., 2015. Expert panel report on the behaviour and environmental impacts of crude oil released into aqueous environments. Ottawa, ON: Royal Society of Canada.; Walker et al., 2019WALKER, T.R., ADEBAMBO, O., DEL AGUILA FEIJOO, M.C., ELHAIMER, E., HOSSAIN, T., EDWARDS, S.J., MORRISON, C.E., ROMO, J., SHARMA, N., TAYLOR, S. and ZOMORODI, S., 2019. Environmental effects of marine transportation. In C. SHEPPARD, ed. World seas: an environmental evaluation (pp. 505-530). Cambridge: Academic Press. http://dx.doi.org/10.1016/B978-0-12-805052-1.00030-9.
http://dx.doi.org/10.1016/B978-0-12-8050...
). Oil spills disrupt marine life and have an impact on the ecosystem years after a spill occurs (Daly et al., 2016DALY, K.L., PASSOW, U., CHANTON, J. and HOLLANDER, D., 2016. Assessing the impacts of oil-associated marine snow formation and sedimentation during and after the Deepwater Horizon oil spill. Anthropocene, vol. 13, pp. 18-33. http://dx.doi.org/10.1016/j.ancene.2016.01.006.
http://dx.doi.org/10.1016/j.ancene.2016....
). Sewage discharges, mechanized fishing boats, cleaning of bilged, tank washing by the huge number of merchant vessels and oil tankers that pass through the Exclusive Economic Zone (EEZ) are main sources of oil pollution in Pakistan (Pastorinho & Pais, 2019PASTORINHO, M. R., & PAIS, R., 2019. Sampling pelagic marine organisms. In: D.-S. PEI and M. JUNAID, eds. Marine ecology: current and future developments. United Arab Emirates: Bentham Science, vol. 1, pp. 12-36.). About 2500 oil tankers transport 33 million tons of crude oil through Pakistani seaports per annuum (Pastorinho & Pais, 2019PASTORINHO, M. R., & PAIS, R., 2019. Sampling pelagic marine organisms. In: D.-S. PEI and M. JUNAID, eds. Marine ecology: current and future developments. United Arab Emirates: Bentham Science, vol. 1, pp. 12-36.). The oil tanker M.T. Tasman Spirit grounded in the channel of Karachi harbor about 1.5 nm from the sea shore on July 27, 2003, while cruising in the curved entry of channel. The vessel was loaded with 67,535 tons of Iranian light crude oil for the Pakistan Refinery Limited in Karachi (Siddiqi and Munshi, 2015SIDDIQI, H.A. and MUNSHI, A.B., 2015. Handbook of oil spill science and technology. In: M. FINGAS, ed. Tasman spirit oil spill at karachi coast, Pakistan (pp. 557-573). Hoboken: John Wiley & Sons, Inc.). In this connection, more than 33,000 tons of oil leaked into the Arabian Sea and 16 km of the coast was contaminated near Karachi in 2003 by the Tasman Spirit (Fingas, 2014FINGAS, M., 2014. Handbook of oil spill science and technology. Hoboken: John Wiley & Sons. http://dx.doi.org/10.1002/9781118989982.
http://dx.doi.org/10.1002/9781118989982...
). Human induced activities and non-tank vessels such as, washing ballast tanks account for 36,000 metric tons of oil entering the oceans globally every year (Fingas, 2013FINGAS, M., 2013. The basics of oil spill cleanup. 3rd ed. UK: Routledge.) . These slow and gradual seeps oil release during an oil spill or extraction accident into the marine environment effect both biotic and abiotic ecosystem to a considerable strength (Blackburn et al., 2014BLACKBURN, M., MAZZACANO, C.A., FALLON, C. and BLACK, S.H., 2014. A review of the impacts of oil spills on marine invertebrates. Portland, Oregon: The Xerces Society.).
Although chemical-physical features play an important role in the process of oil detoxification, the ultimate and complete degradation is mainly accomplished by marine microflora particularly bacteria (Jain et al., 2011JAIN, P., GUPTA, V., GAUR, R., LOWRY, M., JAROLI, D. and CHAUHAN, U.K., 2011. Bioremediation of petroleum oil contaminated soil and water. Research Journal of Environmental Toxicology, vol. 5, no. 1, pp. 1-26. http://dx.doi.org/10.3923/rjet.2011.1.26.
http://dx.doi.org/10.3923/rjet.2011.1.26...
; Santisi et al., 2015SANTISI, S., CAPPELLO, S., CATALFAMO, M., MANCINI, G., HASSANSHAHIAN, M., GENOVESE, L., GIULIANO, L. and YAKIMOV, M.M., 2015. Biodegradation of crude oil by individual strains and a mixed bacterial consortium in simulation of marine environment. Brazilian Journal of Microbiology, vol. 46, no. 2, pp. 377-387.). Bioremediation has been widely used because of its advantages of good effect, easy operation, low cost, rapid degradation rate, and lack of secondary pollution (Zhang et al., 2020ZHANG, C., WU, D. and REN, H., 2020. Bioremediation of oil contaminated soil using agricultural wastes via microbial consortium. Scientific Reports, vol. 10, no. 1, pp. 1-8. http://dx.doi.org/10.1038/s41598-020-66169-5. PMid:32513982.
http://dx.doi.org/10.1038/s41598-020-661...
).
Various microorganisms such as bacteria and Archaea can degrade Petroleum hydrocarbons (Miettinen et al., 2019MIETTINEN, H., BOMBERG, M., NYYSSÖNEN, M., REUNAMO, A., JØRGENSEN, K. S., & VIKMAN, M., 2019. Oil degradation potential of microbial communities in water and sediment of Baltic Sea coastal area. PlosOne, vol. 14, no. 7, pp. e0218834.) from soil and groundwater (Lima et al., 2020LIMA, S., OLIVEIRA, A., GOLIN, R., LOPES, V., CAIXETA, D., LIMA, Z. and MORAIS, E.B., 2020. Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil. Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 80, no. 2, pp. 354-361. http://dx.doi.org/10.1590/1519-6984.208611. PMid:31389483.
http://dx.doi.org/10.1590/1519-6984.2086...
). Among them bacteria like, Pseudomonas fluorescens, P. aeruginosa, Bacillus subtilis, Bacillus sp., Alcaligenes sp., Acinetobacter lwoffi, Flavobacterium sp., Micrococcus roseus, and Corynebacterium sp. were isolated from the polluted stream which could degrade crude oil (Das & Chandran, 2011DAS, N. and CHANDRAN, P., 2011. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnology Research International, vol. 2011, pp. 941810. http://dx.doi.org/10.4061/2011/941810. PMid:21350672.
http://dx.doi.org/10.4061/2011/941810...
). Indigenous oil degrading bacteria in the Arabian Gulf and other aquatic environments occur more frequently in naturally immobilized forms than as free planktonic microorganisms (Radwan, 2008RADWAN, S., 2008. Microbiology of oil-contaminated desert soils and coastal areas in the Arabian Gulf region. In: P. DION and C.S. NAUTIYAL, eds., Microbiology of extreme soils. USA: Springer, pp. 275-298. http://dx.doi.org/10.1007/978-3-540-74231-9_13.
http://dx.doi.org/10.1007/978-3-540-7423...
). Such immobilized oil utilizing bacteria are reported to be associated with wild and farm fish of the Arabian Gulf (Mahmoud et al., 2009MAHMOUD, H., AL-HASAN, R., SORKHOH, N., ELIYAS, M. and RADWAN, S.S., 2009. Attenuation of oil pollutants in the Arabian Gulf water by bacteria naturally associated with live fish. International Biodeterioration & Biodegradation, vol. 63, no. 5, pp. 615-620. http://dx.doi.org/10.1016/j.ibiod.2009.04.005.
http://dx.doi.org/10.1016/j.ibiod.2009.0...
; Radwan et al., 2007RADWAN, S.S., AL-HASAN, R.H., MAHMOUD, H.M. and ELIYAS, M., 2007. Oil‐utilizing bacteria associated with fish from the Arabian Gulf. Journal of Applied Microbiology, vol. 103, no. 6, pp. 2160-2167. http://dx.doi.org/10.1111/j.1365-2672.2007.03454.x. PMid:17953689.
http://dx.doi.org/10.1111/j.1365-2672.20...
). Keeping the context in view, we have characterized the oil degrading bacteria from fish of Indian Ocean at Karachi point. We have further determined the oil degrading potential of the bacteria, which altogether will help in setting ways for further studies on commercial basis.
2. Methods
2.1. Sample collection
A total of 11 fish samples were collected from Indian Ocean near oil port at Karachi Pakistan (Table 1). Fish were aseptically captured using fish hooks and transferred to sterilized polythin bags. The sampling sites were traced using Android App (AndLocation). The traced locations were at 24.798 ̊ Latitude and 66.975 ̊ Longitude with 9m accuracy; 24.784 ̊ Latitude and 66.982 ̊ Longitude with the accuracy of 6m; 24.795 ̊ Latitude and 67.00 ̊ Longitude with 6m accuracy. The fish samples were transported in ice layers to the laboratory in the Department of Biotechnology and Genetic Engineering KUST, Kohat and immediately subject to bacterial isolation. All experimental procedures (capturing, handling and sacrifice of fish) involved in this study were permitted by the Research Ethical Committee of Kohat University of Science and Technology, Kohat.
2.2. Bacterial isolation and counting
Bacteria from the skin, gills and the guts were isolated using sterile surgical blades to scrap off maximum bacterial cells. The bacterial isolation was carried out by standard serial dilution plating technique. Mineral Salt Agar (MSA) having a composition (g/l) of CaCl2. 2H2O, 0.02; K2HPO4, 1.73; KH2PO4, 0.68; NH4NO3, 1; MgSO4. 7H2O, 0.1; NaCl, 4; C6H12O6, 5; FeSO4. 7H2O, 0.03; agar, 20, pH 7.0, (Borah and Yadav, 2014BORAH, D. and YADAV, R.N.S., 2014. Biodegradation of diesel, crude oil, kerosene and used engine oil by a newly isolated Bacillus cereus strain DRDU1 from an automobile engine in liquid culture. Arabian Journal for Science and Engineering, vol. 39, no. 7, pp. 5337-5345. http://dx.doi.org/10.1007/s13369-014-1118-3.
http://dx.doi.org/10.1007/s13369-014-111...
) with crude oil as a sole source of carbon and energy was used. Bacterial colonies were counted by the standard dilution-plate technique.
2.3. Selection of potential oil degrading bacteria
Thirty-six (36) morphologically different colonies were picked from the developed colonies over MSA plates containing crude oil, Potential isolates were selected using gradual increase (0.2%, 0.5% 0.7%, 1%, 2% and 5%) of crude oil concentrations. The selected isolates were subjected to Gram staining and molecular characterization.
2.4. Evaluation of bacterial growth
Bacterial cells were cultured overnight in Lysogeny Broth (LB) media. One (01) ml of the culture was transferred to flasks containing 50 ml of Mineral Salt Medium (MSM) followed by autoclaving for 15 minutes at 121 ºC. Then 0.5 ml of crude oil was added and were incubated over orbital shaker at 150 rpm and at 30 ºC. Three flasks for each bacterial isolate were taken, two of them were inoculated and the 3rd one was kept as a control with no bacterial inoculum having crude oil. Change in turbidity was observed as the measure of growth by taking the optical density (OD) at 600 nm before and after 10 days of inoculation in mineral salt medium.
2.5. Oil degrading potential
Crude and used engine oil (0.5% w/v each) were added to flasks containing 50ml of MS medium. The flasks were inoculated with 1ml overnight culture of Bacillus velezensis (G4A). The flasks were then incubated in shaker at 30 ºC and 150 rpm for 10 days. Control flasks devoid of inoculum, were incubated under the same conditions. After incubation, the residual hydrocarbons were extracted in triplicates using n-hexane as a solvent. The extracted hydrocarbons were analyzed quantitatively using GC-MS using Shimadzu GC-MS QP 2010 PLUS (Japan) with DB5MS column (30 m × 0.25 mm, 0.25μm film thickness), with helium as the carrier gas with a split ratio of 5:50. The oven temperature was programmed at 35 °C for 5 minutes and then increased to 300 °C at 5°C per minute, and held at this temperature for 100 minutes. The compounds were identified by comparison of their mass spectra with the mass spectral libraries.
2.6. DNA extraction and PCR amplification of 16S rRNA gene
The genomic DNA of bacterial isolates was extracted using Gene JET Genomic DNA Purification Kit following manufacturer instructions. Universal primers (forward; 5′-ACGGGCGTGTGTAC-3′ and reverse; 5′-CAGCCGCGGTAATA-3′) were used for the amplification of 16S rRNA gene. Total 25 µl volume of PCR reagent mixture with readymade Master mix 8 µl, PCR water 12 µl, 1 µl of each forward and reverse primer and 3 µl of sample DNA, was prepared. PCR conditions were set as initial denaturation temperature 94 ºC for 5 minutes, denaturation step 94 ºC for 1 minute, annealing 56 ºC for 50 seconds, extension 72 ºC 35 cycles of 50 seconds, and final extension of 1 cycle for 3 minutes. After completion of PCR reaction, the amplified products were confirmed by 2% agarose gel electrophoresis and were sequenced through ABI (Applied Biosystems, Foster City, CA, USA).
2.7. Phylogenetic analysis
The obtained sequences from bacterial isolates were aligned for similarity with the 16S rRNA sequence available in the database (NCBI), using BLAST algorithm using Clustal W. The phylogenetic tree was constructed using Neighbor-Joining method through MEGA 6 software.
3. Results and discussion
Biodegradation is the reliable and environmentally friendly technology for cleaning up oil pollutants from marine environments. Microorganisms specially bacteria play an important role in the degradation of oil pollutants. Indigenous oil degrading bacteria have high potential of biodegradation than freely movable bacteria. Reports has discovered various groups of oil degrading bacteria with their potential from several marine surfaces including water, sediment (Venkateswaran et al., 1991VENKATESWARAN, K., IWABUCHI, T., MATSUI, Y., TOKI, H., HAMADA, E. and TANAKA, H., 1991. Distribution and biodegradation potential of oil-degrading bacteria in North Eastern Japanese coastal waters. FEMS Microbiology Ecology, vol. 86, no. 2, pp. 113-121. http://dx.doi.org/10.1111/j.1574-6968.1991.tb04800.x.
http://dx.doi.org/10.1111/j.1574-6968.19...
), and gut of several fish species (Voverienė et al., 2002VOVERIENĖ, G., MICKĖNIENĖ, L. and ŠYVOKIENĖ, J.S., 2002. Hydrocarbon-degrading bacteria in the digestive tract of fish, their abundance, species composition, and activity. Acta Zoologica Lituanica, vol. 12, no. 3, pp. 333-340. http://dx.doi.org/10.1080/13921657.2002.10512520.
http://dx.doi.org/10.1080/13921657.2002....
). The Indian ocean at Karachi sea port has strong evidences of spilled oil (Pastorinho & Pais, 2019PASTORINHO, M. R., & PAIS, R., 2019. Sampling pelagic marine organisms. In: D.-S. PEI and M. JUNAID, eds. Marine ecology: current and future developments. United Arab Emirates: Bentham Science, vol. 1, pp. 12-36.), therefore the current study collected 11 fish species from Indian ocean near oil port at Karachi. The detail of the collected fish samples was presented in Table 1.
Mineral Salt Agar (MSA) containing 0.2% v/v crude oil was used as a source of carbon and energy. The results showed that fish of Indian ocean accommodated rich bacterial consortium on their skin, gills and guts surfaces. On skin surface, the highest number of bacteria was found of Sea bream, on gill surface, the highest number of bacteria was found of Saddle grunt 1, while on gut surface the highest number of bacteria was found of Indian oil sardine (Table 2). Similar records of oil degrading bacteria associated with wild and farm fish in the neighbor Arabian Gulf were reported in the previous decade (Mahmoud et al., 2009MAHMOUD, H., AL-HASAN, R., SORKHOH, N., ELIYAS, M. and RADWAN, S.S., 2009. Attenuation of oil pollutants in the Arabian Gulf water by bacteria naturally associated with live fish. International Biodeterioration & Biodegradation, vol. 63, no. 5, pp. 615-620. http://dx.doi.org/10.1016/j.ibiod.2009.04.005.
http://dx.doi.org/10.1016/j.ibiod.2009.0...
; Radwan et al., 2007RADWAN, S.S., AL-HASAN, R.H., MAHMOUD, H.M. and ELIYAS, M., 2007. Oil‐utilizing bacteria associated with fish from the Arabian Gulf. Journal of Applied Microbiology, vol. 103, no. 6, pp. 2160-2167. http://dx.doi.org/10.1111/j.1365-2672.2007.03454.x. PMid:17953689.
http://dx.doi.org/10.1111/j.1365-2672.20...
). The digestive tract of mysids, gammarids, fish larvae and adult fish in the Curonian lagoon also contained hydrocarbon degrading bacteria in a similar trend (Šyvokienė et al., 2005ŠYVOKIENĖ, J., MICKĖNIENĖ, L. and JANKAUSKIENĖ, R.J.J., 2005. Hydrocarbon-degrading bacteria in the bacteriocenosis of crustaceans and fish. Acta Zoologica Lituanica, vol. 2, pp. 101-111.). The difference in the strains of bacteria among these studies may be due to varied fish species and/or hydrocarbon type, which in either case need further studies.
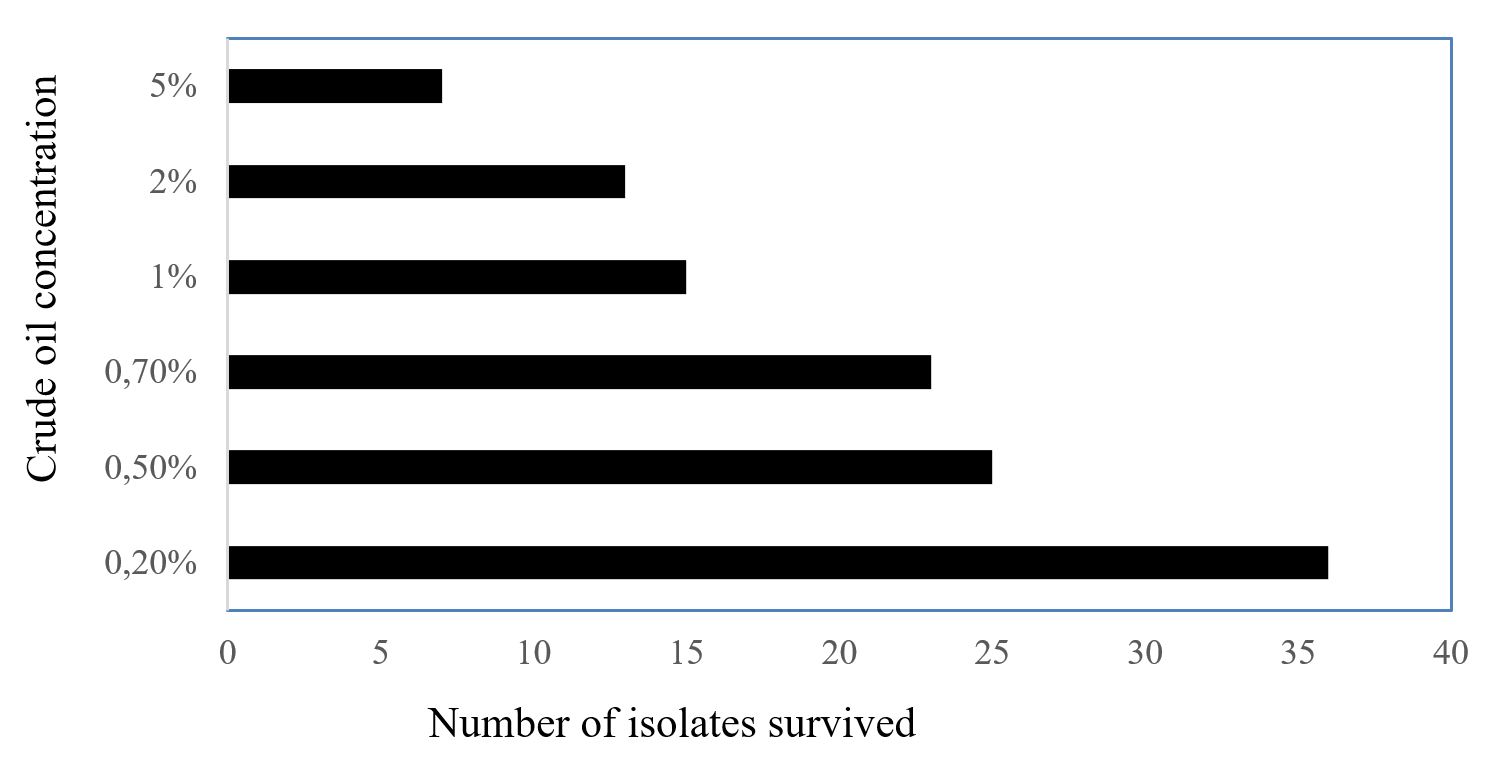
The total 36 bacterial isolates after growing on MSA and nutrient agar media with different concentrations of crude oil were selected based on their growth on different concentration of crude oil. Finally, maximum 7 bacterial isolates were survived on media contained 5% crude oil. The survival of the bacterial strains against the percentage of crude oil was shown in Figure 1. Previous studies also reported various bacterial adaptation to different concentrations of crude oil (Li et al., 2015LI, W., HAN, J.L. and ZHENG, G.A.O., 2015. New findings in effect of different crude oil concentrations on bacterioplankton communities. Microbiology China, vol. 42, no. 5, pp. 826-834.; Sharma and Schiewer, 2016SHARMA, P. and SCHIEWER, S., 2016. Assessment of crude oil biodegradation in arctic seashore sediments: effects of temperature, salinity, and crude oil concentration. Environmental Science and Pollution Research International, vol. 23, no. 15, pp. 14881-14888. http://dx.doi.org/10.1007/s11356-016-6601-9. PMid:27072034.
http://dx.doi.org/10.1007/s11356-016-660...
). (Zhao et al., 2011ZHAO, D., LIU, C., LIU, L., ZHANG, Y., LIU, Q. and WU, W.-M., 2011. Selection of functional consortium for crude oil-contaminated soil remediation. International Biodeterioration & Biodegradation, vol. 65, no. 8, pp. 1244-1248. http://dx.doi.org/10.1016/j.ibiod.2011.07.008.
http://dx.doi.org/10.1016/j.ibiod.2011.0...
) studied selection of functional consortium for the remediation of oil contaminated soil. Taking all the results together, bacterial growth was about similar against various concentration of crude oil.
Bacterial growth against crude oil on MSA media. The Y axis shows the percentage of crude oil concentration while the X axis shows the number of bacterial isolates survived.
Of all the 7 survived isolates, 4 isolates namely G3A, G4A, G2A and L2C that were confirmed with high growth where their growth on MSA and LB media with and/or without 0.5% crude oil was observed after 24 hours (Table 3), (Figure 2 and Figure 3).
Comparison of 24 h bacterial culture using LB broth with/without crude oil and MS broth with/ without crude oil.
Comparison of 24 h bacterial culture using LB broth containing crude oil and MS broth without crude oil.
Comparison of 24 h LB broth culture (without crude oil) and 10-day MSM culture containing 0.5% crude oil.
Gram staining confirmed that two isolates (G3A and G4A) were Gram positive Rods and two isolates (G2A and L2C) were Gram negative Rods.
The 16S rRNA (1450 bp amplicon size) sequencing characterization confirmed that isolates G3A and G4A showed 99% identity with genus Bacillus where as G3A was identical with Bacillus flexus and G4A was identical with Bacillus velezensis. Both bacteria were previously reported as oil degraders (Ren et al., 2012REN, Y.-J., CHEN, M.-M., YUE, Y., WANG, L., WANG, J. and WANG, W-F., 2012. Isolation and degradation properties of heavy oil-degrading strain Bacillus flexus DL1-G. Zhongguo Huanjing Kexue, vol. 32, no. 6, pp. 1080-1086.; Yateem et al., 2011YATEEM, A., AL-SHARRAH, T.J.S. and CONTAMINATION, S., 2011. Enhanced hydrocarbons degradation in the rhizosphere of mangrove plants by a halophilic Bacillus subtilis subtilis strain. Soil and Sediment Contamination: An International Journal, vol. 20, no. 3, pp. 261-273. http://dx.doi.org/10.1080/15320383.2011.560980.
http://dx.doi.org/10.1080/15320383.2011....
). Species of the genus Bacillus are usually scattered in oil contaminated environments due to its low nutritive value (Cybulski et al., 2003CYBULSKI, Z., DZIURLA, E., KACZOREK, E. and OLSZANOWSKI, A., 2003. The influence of emulsifiers on hydrocarbon biodegradation by Pseudomonadacea and Bacillacea strains. Spill Science & Technology Bulletin, vol. 8, no. 5-6, pp. 503-507. http://dx.doi.org/10.1016/S1353-2561(03)00068-9.
http://dx.doi.org/10.1016/S1353-2561(03)...
). Bacillus species have the characteristics of bioremediation (Tian et al., 2018TIAN, X., WANG, X., PENG, S., WANG, Z., ZHOU, R. and TIAN, H., 2018. Isolation, screening, and crude oil degradation characteristics of hydrocarbons-degrading bacteria for treatment of oily wastewater. Water Science and Technology, vol. 78, no. 12, pp. 2626-2638. http://dx.doi.org/10.2166/wst.2019.025. PMid:30767927.
http://dx.doi.org/10.2166/wst.2019.025...
), where marine oil contaminants have been attenuated by Bacillus species (Chikere and Okpokwasili, 2009CHIKERE, C. and OKPOKWASILI, G., 2009. Characterization of hydrocarbon utilizing bacteria in tropical marine sediments. African Journal of Biotechnology, vol. 8, no. 11, pp. 2541-2544.). All these reports suggest that genus Bacillus have high adaptability to several extreme environments. The isolates G2A and L2C were identical with genus Pseudomonas, where the similarity of G2A was 100% with a reference or type strain of Pseudomonas brenneri and similarity of L2C was 99% with reference strain of Pseudomonas azotoformans. Pseudomonas are hydrocarbon degraders (Kostka et al., 2011KOSTKA, J.E., PRAKASH, O., OVERHOLT, W.A., GREEN, S.J., FREYER, G., CANION, A., DELGARDIO, J., NORTON, N., HAZEN, T.C. and HUETTEL, M., 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Applied and Environmental Microbiology, vol. 77, no. 22, pp. 7962-7974. http://dx.doi.org/10.1128/AEM.05402-11. PMid:21948834.
http://dx.doi.org/10.1128/AEM.05402-11...
; Palleroni et al., 2010PALLERONI, N.J., PIEPER, D.H. and MOORE, E.R.B., 2010. Microbiology of hydrocarbon-degrading pseudomonas. In: K.N. TIMMIS, eds. Handbook of hydrocarbon and lipid microbiology. Berlin, Heidelberg: Springer. http://dx.doi.org/10.1007/978-3-540-77587-4_129), that even present in the gut of fish (Voverienė et al., 2002VOVERIENĖ, G., MICKĖNIENĖ, L. and ŠYVOKIENĖ, J.S., 2002. Hydrocarbon-degrading bacteria in the digestive tract of fish, their abundance, species composition, and activity. Acta Zoologica Lituanica, vol. 12, no. 3, pp. 333-340. http://dx.doi.org/10.1080/13921657.2002.10512520.
http://dx.doi.org/10.1080/13921657.2002....
). Various species of Pseudomonas have been widely studied to have oil bio-degradation potential or have close association with oil from various oil contaminated zones (Adebusoye et al., 2007ADEBUSOYE, S.A., ILORI, M.O., AMUND, O.O., TENIOLA, O.D. and OLATOPE, S.O., 2007. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World Journal of Microbiology and Biotechnology, vol. 23, no. 8, pp. 1149-1159.; Bučková et al., 2013BUČKOVÁ, M., PUŠKAROVÁ, A., CHOVANOVÁ, K., KRAKOVÁ, L., FERIANC, P. and PANGALLO, D., 2013. A simple strategy for investigating the diversity and hydrocarbon degradation abilities of cultivable bacteria from contaminated soil. World Journal of Microbiology & Biotechnology, vol. 29, no. 6, pp. 1085-1098. http://dx.doi.org/10.1007/s11274-013-1277-5. PMid:23386320.
http://dx.doi.org/10.1007/s11274-013-127...
; Kaczorek and Olszanowski, 2011KACZOREK, E. and OLSZANOWSKI, A.J.W., 2011. Uptake of hydrocarbon by Pseudomonas fluorescens (P1) and Pseudomonas putida (K1) strains in the presence of surfactants: a cell surface modification. Water, Air, and Soil Pollution, vol. 214, no. 1-4, pp. 451-459. http://dx.doi.org/10.1007/s11270-010-0436-7. PMid:21258434.
http://dx.doi.org/10.1007/s11270-010-043...
; Meliani et al., 2014MELIANI, A., BENSOLTANE, A.J.J.P. and BIOTECHNOLOGY, E., 2014. Enhancement of hydrocarbons degradation by use of Pseudomonas biosurfactants and biofilms. Journal of Petroleum & Environmental Biotechnology, vol. 5, no. 1, pp. 1. http://dx.doi.org/10.4172/2157-7463.1000168.
http://dx.doi.org/10.4172/2157-7463.1000...
; Wald et al., 2015WALD, J., HROUDOVA, M., JANSA, J., VRCHOTOVA, B., MACEK, T. and UHLIK, O., 2015. Pseudomonads rule degradation of polyaromatic hydrocarbons in aerated sediment. Frontiers in Microbiology, vol. 6, pp. 1268. http://dx.doi.org/10.3389/fmicb.2015.01268. PMid:26635740.
http://dx.doi.org/10.3389/fmicb.2015.012...
). In order to further characterize and assess the position of the sequence of the isolated strains with the sequence of their sister strains published on NCBI, phylogenetic tree was created by Neighbor-joining method for the purpose as describe by Tamur et al. (Tamura et al., 2011TAMURA, K., PETERSON, D., PETERSON, N., STECHER, G., NEI, M. and KUMAR, S., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, vol. 28, no. 10, pp. 2731-2739. http://dx.doi.org/10.1093/molbev/msr121. PMid:21546353.
http://dx.doi.org/10.1093/molbev/msr121...
) and Kumar et al. (Kumar et al., 2018KUMAR, S., STECHER, G., LI, M., KNYAZ, C. and TAMURA, K., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, vol. 35, no. 6, pp. 1547-1549. http://dx.doi.org/10.1093/molbev/msy096. PMid:29722887.
http://dx.doi.org/10.1093/molbev/msy096...
) (Figure 4).
Phylogenetic tree showing position of 4 isolated strains. The optimal tree with the sum of branch length = 0.20627849 is shown. The nucleotide sequence used for finding evolutionary relationship was ~ 950 bp. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. This analysis involved 16 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 844 positions in the final dataset.
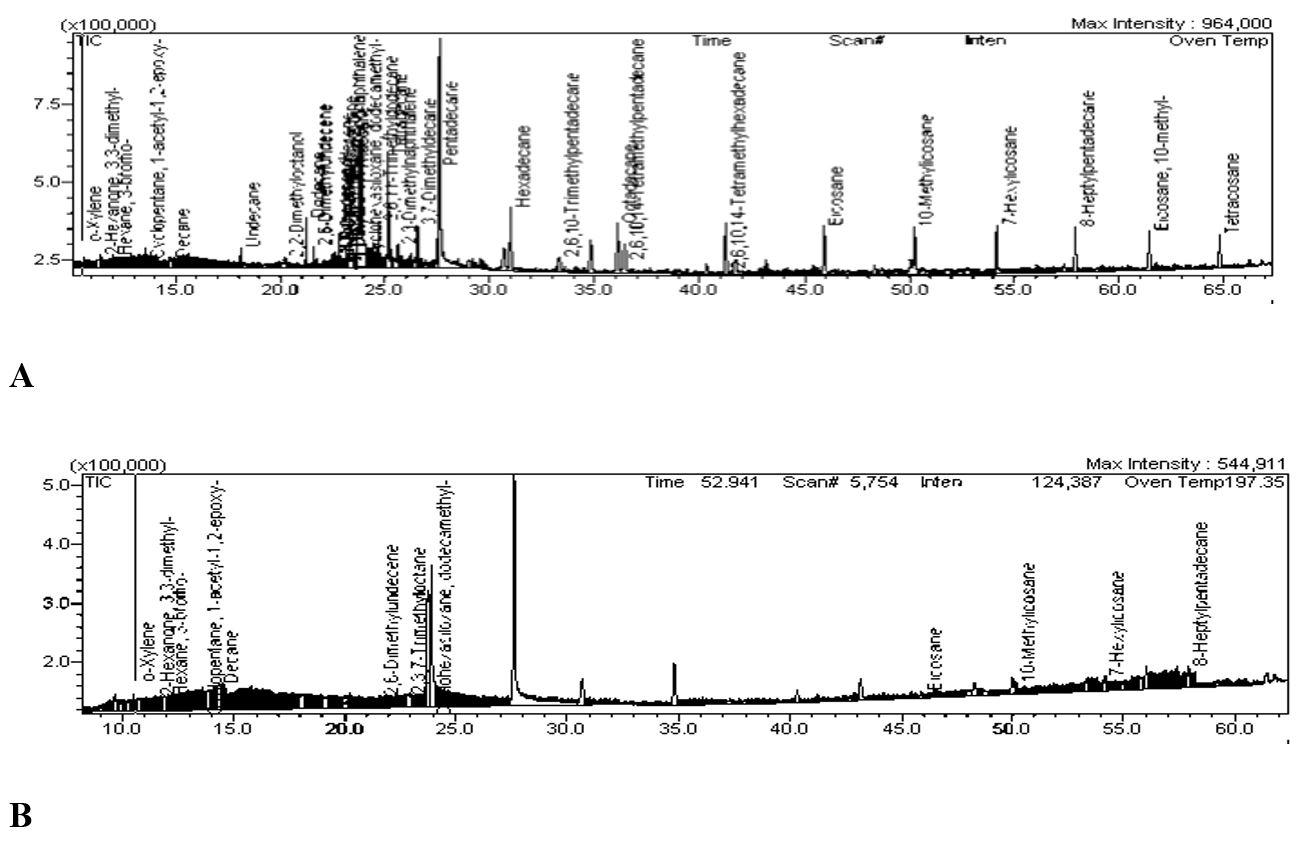
Since the strain B. velezensis (G4A) showed high growth with high OD value than other strains after 10 days incubation (Table 4). Therefore, we have selected this strain for further biodegradation of crude oil and used engine oil. The strain B. velezensis was inoculated in media containing crude oil and used engine oil respectively and the total petroleum hydrocarbons (TPHs) were extracted from the oil-amended treatments after 10 days. Oil degradation potential was determined against the results of bacterial treatments to those of the uninoculated control. In case of crude oil there were total 42 components present in control set while 41 in treated set. Only one component is eliminated and the concentration of remaining is reduced (Figure 5).
Optical density of the isolates in Mineral Salt broth containing crude oil after 10-day incubation in shaker
(A) GC-MS spectra Control Crude oil (No inoculation); (B) GC-MS spectra Sample crude oil (Inoculated).
In case of used engine oil there were total 42 components in control set while 16 components in treated set. In this case the bacteria degraded 26 components of used engine oil were eliminated and the concentration of some components were amended (Figure 6). Taken together, the result suggests that the selected strain has potential to degrade used engine oil better than aged crude oil. It is evident that Bacillus species has high degradation potential for used engine oil lower (28%) than crude oil (84%) (Raju et al., 2017RAJU, M.N., LEO, R., HERMINIA, S.S., MORÁN, R.E., VENKATESWARLU, K. and LAURA, S., 2017. Biodegradation of Diesel, Crude Oil and Spent Lubricating Oil by Soil Isolates of Bacillus spp. Bulletin of Environmental Contamination and Toxicology, vol. 98, no. 5, pp. 698-705. http://dx.doi.org/10.1007/s00128-017-2039-0. PMid:28210752.
http://dx.doi.org/10.1007/s00128-017-203...
). Borah and Yadav (2014)BORAH, D. and YADAV, R.N.S., 2014. Biodegradation of diesel, crude oil, kerosene and used engine oil by a newly isolated Bacillus cereus strain DRDU1 from an automobile engine in liquid culture. Arabian Journal for Science and Engineering, vol. 39, no. 7, pp. 5337-5345. http://dx.doi.org/10.1007/s13369-014-1118-3.
http://dx.doi.org/10.1007/s13369-014-111...
confirmed that Bacillus species degrade used engine oil, crude oil, diesels oil and kerosene as 42%, 57%, 67% and 72% respectively. Our results deviate the previous studies as they reported low degradation of used engine oil than crude oil by Bacillus. It may be due to the conditions of hydrocarbon, culturing conditions and species of bacteria. In our experiment we used aged crude oil that can degrades slower than other hydrocarbons by bacteria (Kang et al., 2009KANG, Y.-S., PARK, Y.-J., JUNG, J.-J. and PARK, W., 2009. Inhibitory effect of aged petroleum hydrocarbons on the survival of inoculated microorganism in a crude-oil-contaminated site. Journal of Microbiology and Biotechnology, vol. 19, no. 12, pp. 1672-1678. http://dx.doi.org/10.4014/jmb.0903.03001. PMid:20075636.
http://dx.doi.org/10.4014/jmb.0903.03001...
). However, further studies are needed for quantification of individual oil/ hydrocarbon degrading potential of this bacteria and characterization of its potential enzymes for this process.
(A) GC-MS spectra Control used Engine oil (No inoculation). B. GC-MS spectra Used Engine oil sample (inoculated).
4. Conclusion
This study isolated and characterized fish associated oil degrading bacteria related to B. velezensis, B. flexus, P. brenneri and P. azotofarmens from Indian ocean near Karachi sea port. We further confirmed that Bacillus velezensis have high growth potential on oil containing media. The GC-MS technology showed that B. velezensis have degraded major components of used engine oil and a component of crude oil. This report confirmed that oil degrading bacteria residing fish of Indian ocean has the potential to degrade oil that may need further studies for the purpose of bioremediation in future.
Acknowledgements
The authors are grateful to the Higher Education Commission of Pakistan for financial support and the Department of Biotechnology and Genetic Engineering, Kohat University of Science and Technology for providing laboratory facilities to carry out research work. This work was also supported by Research Assistance Program (2019) of the Incheon National University.
References
- ADEBUSOYE, S.A., ILORI, M.O., AMUND, O.O., TENIOLA, O.D. and OLATOPE, S.O., 2007. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World Journal of Microbiology and Biotechnology, vol. 23, no. 8, pp. 1149-1159.
- BLACKBURN, M., MAZZACANO, C.A., FALLON, C. and BLACK, S.H., 2014. A review of the impacts of oil spills on marine invertebrates Portland, Oregon: The Xerces Society.
- BORAH, D. and YADAV, R.N.S., 2014. Biodegradation of diesel, crude oil, kerosene and used engine oil by a newly isolated Bacillus cereus strain DRDU1 from an automobile engine in liquid culture. Arabian Journal for Science and Engineering, vol. 39, no. 7, pp. 5337-5345. http://dx.doi.org/10.1007/s13369-014-1118-3
» http://dx.doi.org/10.1007/s13369-014-1118-3 - BUČKOVÁ, M., PUŠKAROVÁ, A., CHOVANOVÁ, K., KRAKOVÁ, L., FERIANC, P. and PANGALLO, D., 2013. A simple strategy for investigating the diversity and hydrocarbon degradation abilities of cultivable bacteria from contaminated soil. World Journal of Microbiology & Biotechnology, vol. 29, no. 6, pp. 1085-1098. http://dx.doi.org/10.1007/s11274-013-1277-5 PMid:23386320.
» http://dx.doi.org/10.1007/s11274-013-1277-5 - CHIKERE, C. and OKPOKWASILI, G., 2009. Characterization of hydrocarbon utilizing bacteria in tropical marine sediments. African Journal of Biotechnology, vol. 8, no. 11, pp. 2541-2544.
- CYBULSKI, Z., DZIURLA, E., KACZOREK, E. and OLSZANOWSKI, A., 2003. The influence of emulsifiers on hydrocarbon biodegradation by Pseudomonadacea and Bacillacea strains. Spill Science & Technology Bulletin, vol. 8, no. 5-6, pp. 503-507. http://dx.doi.org/10.1016/S1353-2561(03)00068-9
» http://dx.doi.org/10.1016/S1353-2561(03)00068-9 - DALY, K.L., PASSOW, U., CHANTON, J. and HOLLANDER, D., 2016. Assessing the impacts of oil-associated marine snow formation and sedimentation during and after the Deepwater Horizon oil spill. Anthropocene, vol. 13, pp. 18-33. http://dx.doi.org/10.1016/j.ancene.2016.01.006
» http://dx.doi.org/10.1016/j.ancene.2016.01.006 - DAS, N. and CHANDRAN, P., 2011. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnology Research International, vol. 2011, pp. 941810. http://dx.doi.org/10.4061/2011/941810 PMid:21350672.
» http://dx.doi.org/10.4061/2011/941810 - FINGAS, M., 2013. The basics of oil spill cleanup 3rd ed. UK: Routledge.
- FINGAS, M., 2014. Handbook of oil spill science and technology Hoboken: John Wiley & Sons. http://dx.doi.org/10.1002/9781118989982
» http://dx.doi.org/10.1002/9781118989982 - JAIN, P., GUPTA, V., GAUR, R., LOWRY, M., JAROLI, D. and CHAUHAN, U.K., 2011. Bioremediation of petroleum oil contaminated soil and water. Research Journal of Environmental Toxicology, vol. 5, no. 1, pp. 1-26. http://dx.doi.org/10.3923/rjet.2011.1.26
» http://dx.doi.org/10.3923/rjet.2011.1.26 - KACZOREK, E. and OLSZANOWSKI, A.J.W., 2011. Uptake of hydrocarbon by Pseudomonas fluorescens (P1) and Pseudomonas putida (K1) strains in the presence of surfactants: a cell surface modification. Water, Air, and Soil Pollution, vol. 214, no. 1-4, pp. 451-459. http://dx.doi.org/10.1007/s11270-010-0436-7 PMid:21258434.
» http://dx.doi.org/10.1007/s11270-010-0436-7 - KANG, Y.-S., PARK, Y.-J., JUNG, J.-J. and PARK, W., 2009. Inhibitory effect of aged petroleum hydrocarbons on the survival of inoculated microorganism in a crude-oil-contaminated site. Journal of Microbiology and Biotechnology, vol. 19, no. 12, pp. 1672-1678. http://dx.doi.org/10.4014/jmb.0903.03001 PMid:20075636.
» http://dx.doi.org/10.4014/jmb.0903.03001 - KOSTKA, J.E., PRAKASH, O., OVERHOLT, W.A., GREEN, S.J., FREYER, G., CANION, A., DELGARDIO, J., NORTON, N., HAZEN, T.C. and HUETTEL, M., 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Applied and Environmental Microbiology, vol. 77, no. 22, pp. 7962-7974. http://dx.doi.org/10.1128/AEM.05402-11 PMid:21948834.
» http://dx.doi.org/10.1128/AEM.05402-11 - KUMAR, S., STECHER, G., LI, M., KNYAZ, C. and TAMURA, K., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, vol. 35, no. 6, pp. 1547-1549. http://dx.doi.org/10.1093/molbev/msy096 PMid:29722887.
» http://dx.doi.org/10.1093/molbev/msy096 - LEE, K., BOUFADEL, M., CHEN, B., FOGHT, J., HODSON, P., SWANSON, S. and VENOSA, A., 2015. Expert panel report on the behaviour and environmental impacts of crude oil released into aqueous environments Ottawa, ON: Royal Society of Canada.
- LI, W., HAN, J.L. and ZHENG, G.A.O., 2015. New findings in effect of different crude oil concentrations on bacterioplankton communities. Microbiology China, vol. 42, no. 5, pp. 826-834.
- LIMA, S., OLIVEIRA, A., GOLIN, R., LOPES, V., CAIXETA, D., LIMA, Z. and MORAIS, E.B., 2020. Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil. Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 80, no. 2, pp. 354-361. http://dx.doi.org/10.1590/1519-6984.208611 PMid:31389483.
» http://dx.doi.org/10.1590/1519-6984.208611 - MAHMOUD, H., AL-HASAN, R., SORKHOH, N., ELIYAS, M. and RADWAN, S.S., 2009. Attenuation of oil pollutants in the Arabian Gulf water by bacteria naturally associated with live fish. International Biodeterioration & Biodegradation, vol. 63, no. 5, pp. 615-620. http://dx.doi.org/10.1016/j.ibiod.2009.04.005
» http://dx.doi.org/10.1016/j.ibiod.2009.04.005 - MELIANI, A., BENSOLTANE, A.J.J.P. and BIOTECHNOLOGY, E., 2014. Enhancement of hydrocarbons degradation by use of Pseudomonas biosurfactants and biofilms. Journal of Petroleum & Environmental Biotechnology, vol. 5, no. 1, pp. 1. http://dx.doi.org/10.4172/2157-7463.1000168
» http://dx.doi.org/10.4172/2157-7463.1000168 - MIETTINEN, H., BOMBERG, M., NYYSSÖNEN, M., REUNAMO, A., JØRGENSEN, K. S., & VIKMAN, M., 2019. Oil degradation potential of microbial communities in water and sediment of Baltic Sea coastal area. PlosOne, vol. 14, no. 7, pp. e0218834.
- PALLERONI, N.J., PIEPER, D.H. and MOORE, E.R.B., 2010. Microbiology of hydrocarbon-degrading pseudomonas. In: K.N. TIMMIS, eds. Handbook of hydrocarbon and lipid microbiology Berlin, Heidelberg: Springer. http://dx.doi.org/10.1007/978-3-540-77587-4_129
- PASTORINHO, M. R., & PAIS, R., 2019. Sampling pelagic marine organisms. In: D.-S. PEI and M. JUNAID, eds. Marine ecology: current and future developments United Arab Emirates: Bentham Science, vol. 1, pp. 12-36.
- RADWAN, S., 2008. Microbiology of oil-contaminated desert soils and coastal areas in the Arabian Gulf region. In: P. DION and C.S. NAUTIYAL, eds., Microbiology of extreme soils USA: Springer, pp. 275-298. http://dx.doi.org/10.1007/978-3-540-74231-9_13
» http://dx.doi.org/10.1007/978-3-540-74231-9_13 - RADWAN, S.S., AL-HASAN, R.H., MAHMOUD, H.M. and ELIYAS, M., 2007. Oil‐utilizing bacteria associated with fish from the Arabian Gulf. Journal of Applied Microbiology, vol. 103, no. 6, pp. 2160-2167. http://dx.doi.org/10.1111/j.1365-2672.2007.03454.x PMid:17953689.
» http://dx.doi.org/10.1111/j.1365-2672.2007.03454.x - RAJU, M.N., LEO, R., HERMINIA, S.S., MORÁN, R.E., VENKATESWARLU, K. and LAURA, S., 2017. Biodegradation of Diesel, Crude Oil and Spent Lubricating Oil by Soil Isolates of Bacillus spp. Bulletin of Environmental Contamination and Toxicology, vol. 98, no. 5, pp. 698-705. http://dx.doi.org/10.1007/s00128-017-2039-0 PMid:28210752.
» http://dx.doi.org/10.1007/s00128-017-2039-0 - REN, Y.-J., CHEN, M.-M., YUE, Y., WANG, L., WANG, J. and WANG, W-F., 2012. Isolation and degradation properties of heavy oil-degrading strain Bacillus flexus DL1-G. Zhongguo Huanjing Kexue, vol. 32, no. 6, pp. 1080-1086.
- SANTISI, S., CAPPELLO, S., CATALFAMO, M., MANCINI, G., HASSANSHAHIAN, M., GENOVESE, L., GIULIANO, L. and YAKIMOV, M.M., 2015. Biodegradation of crude oil by individual strains and a mixed bacterial consortium in simulation of marine environment. Brazilian Journal of Microbiology, vol. 46, no. 2, pp. 377-387.
- SHARMA, P. and SCHIEWER, S., 2016. Assessment of crude oil biodegradation in arctic seashore sediments: effects of temperature, salinity, and crude oil concentration. Environmental Science and Pollution Research International, vol. 23, no. 15, pp. 14881-14888. http://dx.doi.org/10.1007/s11356-016-6601-9 PMid:27072034.
» http://dx.doi.org/10.1007/s11356-016-6601-9 - SIDDIQI, H.A. and MUNSHI, A.B., 2015. Handbook of oil spill science and technology. In: M. FINGAS, ed. Tasman spirit oil spill at karachi coast, Pakistan (pp. 557-573). Hoboken: John Wiley & Sons, Inc.
- ŠYVOKIENĖ, J., MICKĖNIENĖ, L. and JANKAUSKIENĖ, R.J.J., 2005. Hydrocarbon-degrading bacteria in the bacteriocenosis of crustaceans and fish. Acta Zoologica Lituanica, vol. 2, pp. 101-111.
- TAMURA, K., PETERSON, D., PETERSON, N., STECHER, G., NEI, M. and KUMAR, S., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, vol. 28, no. 10, pp. 2731-2739. http://dx.doi.org/10.1093/molbev/msr121 PMid:21546353.
» http://dx.doi.org/10.1093/molbev/msr121 - TIAN, X., WANG, X., PENG, S., WANG, Z., ZHOU, R. and TIAN, H., 2018. Isolation, screening, and crude oil degradation characteristics of hydrocarbons-degrading bacteria for treatment of oily wastewater. Water Science and Technology, vol. 78, no. 12, pp. 2626-2638. http://dx.doi.org/10.2166/wst.2019.025 PMid:30767927.
» http://dx.doi.org/10.2166/wst.2019.025 - VENKATESWARAN, K., IWABUCHI, T., MATSUI, Y., TOKI, H., HAMADA, E. and TANAKA, H., 1991. Distribution and biodegradation potential of oil-degrading bacteria in North Eastern Japanese coastal waters. FEMS Microbiology Ecology, vol. 86, no. 2, pp. 113-121. http://dx.doi.org/10.1111/j.1574-6968.1991.tb04800.x
» http://dx.doi.org/10.1111/j.1574-6968.1991.tb04800.x - VOVERIENĖ, G., MICKĖNIENĖ, L. and ŠYVOKIENĖ, J.S., 2002. Hydrocarbon-degrading bacteria in the digestive tract of fish, their abundance, species composition, and activity. Acta Zoologica Lituanica, vol. 12, no. 3, pp. 333-340. http://dx.doi.org/10.1080/13921657.2002.10512520
» http://dx.doi.org/10.1080/13921657.2002.10512520 - WALD, J., HROUDOVA, M., JANSA, J., VRCHOTOVA, B., MACEK, T. and UHLIK, O., 2015. Pseudomonads rule degradation of polyaromatic hydrocarbons in aerated sediment. Frontiers in Microbiology, vol. 6, pp. 1268. http://dx.doi.org/10.3389/fmicb.2015.01268 PMid:26635740.
» http://dx.doi.org/10.3389/fmicb.2015.01268 - WALKER, T.R., ADEBAMBO, O., DEL AGUILA FEIJOO, M.C., ELHAIMER, E., HOSSAIN, T., EDWARDS, S.J., MORRISON, C.E., ROMO, J., SHARMA, N., TAYLOR, S. and ZOMORODI, S., 2019. Environmental effects of marine transportation. In C. SHEPPARD, ed. World seas: an environmental evaluation (pp. 505-530). Cambridge: Academic Press. http://dx.doi.org/10.1016/B978-0-12-805052-1.00030-9
» http://dx.doi.org/10.1016/B978-0-12-805052-1.00030-9 - YATEEM, A., AL-SHARRAH, T.J.S. and CONTAMINATION, S., 2011. Enhanced hydrocarbons degradation in the rhizosphere of mangrove plants by a halophilic Bacillus subtilis subtilis strain. Soil and Sediment Contamination: An International Journal, vol. 20, no. 3, pp. 261-273. http://dx.doi.org/10.1080/15320383.2011.560980
» http://dx.doi.org/10.1080/15320383.2011.560980 - ZHANG, C., WU, D. and REN, H., 2020. Bioremediation of oil contaminated soil using agricultural wastes via microbial consortium. Scientific Reports, vol. 10, no. 1, pp. 1-8. http://dx.doi.org/10.1038/s41598-020-66169-5 PMid:32513982.
» http://dx.doi.org/10.1038/s41598-020-66169-5 - ZHAO, D., LIU, C., LIU, L., ZHANG, Y., LIU, Q. and WU, W.-M., 2011. Selection of functional consortium for crude oil-contaminated soil remediation. International Biodeterioration & Biodegradation, vol. 65, no. 8, pp. 1244-1248. http://dx.doi.org/10.1016/j.ibiod.2011.07.008
» http://dx.doi.org/10.1016/j.ibiod.2011.07.008
Publication Dates
-
Publication in this collection
28 June 2021 -
Date of issue
2022
History
-
Received
19 Oct 2020 -
Accepted
03 Dec 2020